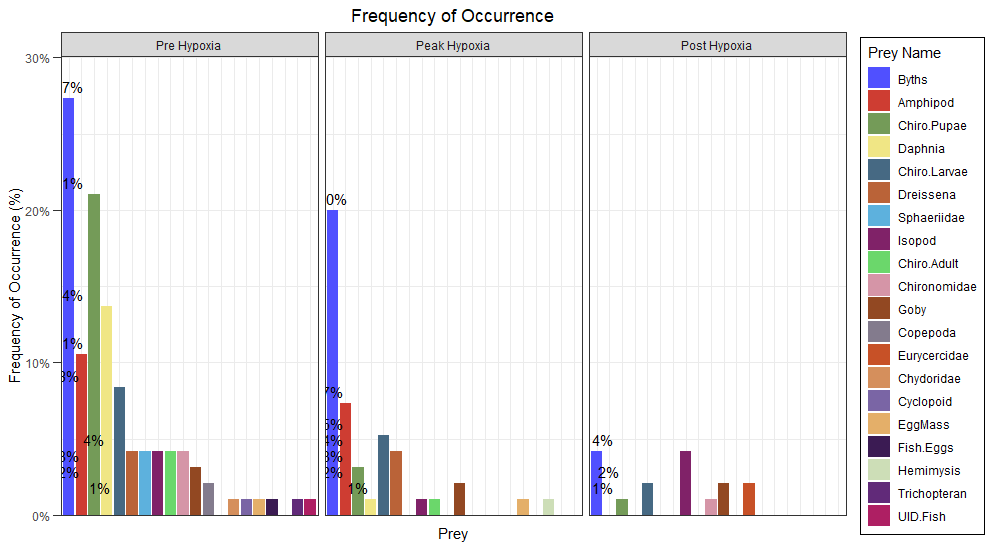
I am trying to add percentage labels to a bar plot using ggplot geom_text but the labels are not in the correct positions. I would like them placed on top of each bar. This question is similar to something I asked about a month ago 
As shown, I would like the bars to have percentage at the top. Bars (prey) that have the same value, should only have it printed once to avoid overcrowding (this part is what I haven't been able to properly adjust).
This is my data when attempting to plot something similar but faceting by group (Season):
structure(list(prey_name = structure(c(2L, 2L, 1L, 1L, 1L, 9L,
9L, 5L, 5L, 5L, 3L, 3L, 3L, 10L, 10L, 14L, 12L, 15L, 4L, 4L,
6L, 6L, 16L, 16L, 13L, 17L, 11L, 11L, 11L, 18L, 8L, 8L, 8L, 7L,
19L, 20L), levels = c("Byths", "Amphipod", "Chiro.Pupae", "Daphnia",
"Chiro.Larvae", "Dreissena", "Sphaeriidae", "Isopod", "Chiro.Adult",
"Chironomidae", "Goby", "Copepoda", "Eurycercidae", "Chydoridae",
"Cyclopoid", "EggMass", "Fish.Eggs", "Hemimysis", "Trichopteran",
"UID.Fish"), class = "factor", scores = structure(c(Amphipod = -0.0894736842105263,
Byths = -0.171929824561404, Chiro.Adult = -0.0263157894736842,
Chiro.Larvae = -0.0526315789473684, Chiro.Pupae = -0.0842105263157895,
Chironomidae = -0.0263157894736842, Chydoridae = -0.0105263157894737,
Copepoda = -0.0210526315789474, Cyclopoid = -0.0105263157894737,
Daphnia = -0.0736842105263158, Dreissena = -0.0421052631578947,
EggMass = -0.0105263157894737, Eurycercidae = -0.0210526315789474,
Fish.Eggs = -0.0105263157894737, Goby = -0.0245614035087719,
Hemimysis = -0.0105263157894737, Isopod = -0.0315789473684211,
Sphaeriidae = -0.0421052631578947, Trichopteran = -0.0105263157894737,
UID.Fish = -0.0105263157894737), dim = 20L, dimnames = list(c("Amphipod",
"Byths", "Chiro.Adult", "Chiro.Larvae", "Chiro.Pupae", "Chironomidae",
"Chydoridae", "Copepoda", "Cyclopoid", "Daphnia", "Dreissena",
"EggMass", "Eurycercidae", "Fish.Eggs", "Goby", "Hemimysis",
"Isopod", "Sphaeriidae", "Trichopteran", "UID.Fish")))), Season = structure(c(1L,
2L, 1L, 2L, 3L, 1L, 2L, 1L, 2L, 3L, 1L, 2L, 3L, 1L, 3L, 1L, 1L,
1L, 1L, 2L, 1L, 2L, 1L, 2L, 3L, 1L, 1L, 2L, 3L, 2L, 1L, 2L, 3L,
1L, 1L, 1L), levels = c("Pre Hypoxia", "Peak Hypoxia", "Post Hypoxia"
), class = "factor"), Fi = c(0.105263157894737, 0.0736842105263158,
0.273684210526316, 0.2, 0.0421052631578947, 0.0421052631578947,
0.0105263157894737, 0.0842105263157895, 0.0526315789473684, 0.0210526315789474,
0.210526315789474, 0.0315789473684211, 0.0105263157894737, 0.0421052631578947,
0.0105263157894737, 0.0105263157894737, 0.0210526315789474, 0.0105263157894737,
0.136842105263158, 0.0105263157894737, 0.0421052631578947, 0.0421052631578947,
0.0105263157894737, 0.0105263157894737, 0.0210526315789474, 0.0105263157894737,
0.0315789473684211, 0.0210526315789474, 0.0210526315789474, 0.0105263157894737,
0.0421052631578947, 0.0105263157894737, 0.0421052631578947, 0.0421052631578947,
0.0105263157894737, 0.0105263157894737)), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"), row.names = c(NA, -36L), groups = structure(list(
prey_name = structure(1:20, levels = c("Byths", "Amphipod",
"Chiro.Pupae", "Daphnia", "Chiro.Larvae", "Dreissena", "Sphaeriidae",
"Isopod", "Chiro.Adult", "Chironomidae", "Goby", "Copepoda",
"Eurycercidae", "Chydoridae", "Cyclopoid", "EggMass", "Fish.Eggs",
"Hemimysis", "Trichopteran", "UID.Fish"), scores = structure(c(Amphipod = -0.0894736842105263,
Byths = -0.171929824561404, Chiro.Adult = -0.0263157894736842,
Chiro.Larvae = -0.0526315789473684, Chiro.Pupae = -0.0842105263157895,
Chironomidae = -0.0263157894736842, Chydoridae = -0.0105263157894737,
Copepoda = -0.0210526315789474, Cyclopoid = -0.0105263157894737,
Daphnia = -0.0736842105263158, Dreissena = -0.0421052631578947,
EggMass = -0.0105263157894737, Eurycercidae = -0.0210526315789474,
Fish.Eggs = -0.0105263157894737, Goby = -0.0245614035087719,
Hemimysis = -0.0105263157894737, Isopod = -0.0315789473684211,
Sphaeriidae = -0.0421052631578947, Trichopteran = -0.0105263157894737,
UID.Fish = -0.0105263157894737), dim = 20L, dimnames = list(
c("Amphipod", "Byths", "Chiro.Adult", "Chiro.Larvae",
"Chiro.Pupae", "Chironomidae", "Chydoridae", "Copepoda",
"Cyclopoid", "Daphnia", "Dreissena", "EggMass", "Eurycercidae",
"Fish.Eggs", "Goby", "Hemimysis", "Isopod", "Sphaeriidae",
"Trichopteran", "UID.Fish"))), class = "factor"), .rows = structure(list(
3:5, 1:2, 11:13, 19:20, 8:10, 21:22, 34L, 31:33, 6:7,
14:15, 27:29, 17L, 25L, 16L, 18L, 23:24, 26L, 30L, 35L,
36L), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -20L), .drop = TRUE))
For my new plot, I tried:
ggplot(FO_adult1, aes(x=reorder(prey_name, -Fi), Fi, fill=prey_name))
geom_bar(stat = "identity")
facet_wrap(~ Season)
geom_text(data = FO_adult1 %>%
mutate(label = scales::percent(round(Fi, digits=2)),
prey_num = as.numeric(reorder(prey_name, -Fi))) %>%
group_by(label, Season) %>%
summarize(n = n(),
label = first(label),
Fi = first(Fi),
prey_num = first(prey_num),
prey_name = first(prey_name)),
aes(x = prey_num (n - 1)/2, y = Fi, label = label),
vjust = -0.5
)
ggtitle("Frequency of Occurrence")
labs(x="Prey", fill = "Prey Name", y = "Frequency of Occurrence (%)")
scale_fill_igv(palette = "default")
theme_bw()
theme(legend.position = "right",
plot.title = element_text(hjust=0.5),
legend.background = element_rect(fill = "white", color = 1),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.length = unit(0.2,"cm"))
scale_y_continuous(expand = expansion(mult = c(0,0.1)), labels=scales::percent)
I tried using the code I used for my original plot but I am not sure what I need to tweak. My biggest hurdle is getting only one percentage label for prey with the same percentage. I would also like to get rid of the empty space/empty bars and plot the groups in descending order if possible (but this is not vital since that would cause each season to be plotted in a different order - but I would like to see what it looks like).
Thank you!
CodePudding user response:
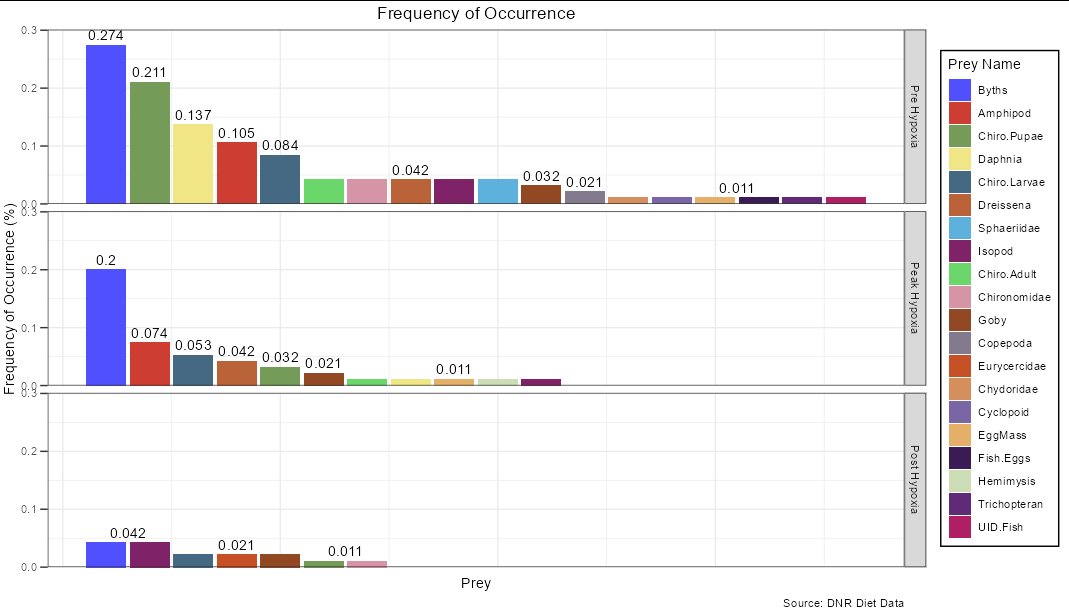
This is a bit trickier, but again can be handled using data manipulation. You need to work out the positions after first grouping by Season
ggplot(FO_adult %>%
group_by(Season) %>%
arrange(-Fi, by_group = TRUE) %>%
mutate(prey_no = seq_along(Fi)) %>%
ungroup(),
aes(x = prey_no, Fi, fill = reorder(prey_name, -Fi)))
geom_col()
facet_grid(Season~., scales = 'free_x')
geom_text(data = . %>%
mutate(label = round(Fi, digits = 3)) %>%
group_by(label, Season) %>%
summarize(n = n(),
label = first(label),
Fi = first(Fi),
prey_no = first(prey_no),
prey_name = first(prey_name)),
aes(x = prey_no (n - 1)/2, y = Fi, label = label),
vjust = -0.5,
check_overlap = TRUE)
ggtitle("Frequency of Occurrence")
labs(x="Prey", fill = "Prey Name", y = "Frequency of Occurrence (%)",
caption = "Source: DNR Diet Data")
ggsci::scale_fill_igv(palette = "default")
theme_bw()
theme(legend.position = "right",
plot.title = element_text(hjust=0.5),
legend.background = element_rect(fill = "white", color = 1),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.length = unit(0.2,"cm"))
scale_y_continuous(expand = expansion(mult = c(0,0.1)))