I am trying to plot a correlation matrix and I am getting this error.
corrplot 0.92 loaded
Error:
! Can't combine ï.. and school .
Backtrace:
- corrplot::corrplot(d1)
- tibble:::
[.tbl_df(corr, !is.na(corr)) - tibble:::tbl_subset_matrix(x, j, j_arg)
- vctrs::vec_c(!!!values, .name_spec = ~.x)
- vctrs
<fn>() - vctrs::vec_default_ptype2(...)
- vctrs::stop_incompatible_type(...)
- vctrs:::stop_incompatible(...)
- vctrs:::stop_vctrs(...)
data_complete <- data[!(data$positivity_rate=="N/A"),]
d1 <- data_complete %>%
group_by(conference, state, school)
d1$positivity_rate <- as.numeric(d1$positivity_rate)
corrplot(d1)
structure(list(ï.. = c(NA_integer_, NA_integer_, NA_integer_,
NA_integer_, NA_integer_, NA_integer_), school = c("Air Force",
"Air Force", "Air Force", "Air Force", "Air Force", "Air Force"
), state = c("Colorado", "Colorado", "Colorado", "Colorado",
"Colorado", "Colorado"), conference = c("Mountain West", "Mountain West",
"Mountain West", "Mountain West", "Mountain West", "Mountain West"
), win_pct = c(0.5, 0.5, 0.5, 0.5, 0.5, 0.5), Wins = c(3L, 3L,
3L, 3L, 3L, 3L), Losses = c(3L, 3L, 3L, 3L, 3L, 3L), pred_ranking = c("",
"", "", "", "", ""), actual_ranking = c("", "", "", "", "", ""
), week = 1:6, positivity_rate = c("0.032", "0.114", "0.034",
"0.039", "0.066", "0.092"), weekly_newcases_per100k = c("86",
"152.8", "190.9", "486.6", "773.8", "1232"), return_to_game_after_positive_test_days = c(NA_integer_,
NA_integer_, NA_integer_, NA_integer_, NA_integer_, NA_integer_
), X7_day_positivity_rate_influencing_games_and_practices = c(NA_integer_,
NA_integer_, NA_integer_, NA_integer_, NA_integer_, NA_integer_
), req_testing_practices = c(NA_integer_, NA_integer_, NA_integer_,
NA_integer_, NA_integer_, NA_integer_), required_72_hr_testing_before_a_game = c(NA_integer_,
NA_integer_, NA_integer_, NA_integer_, NA_integer_, NA_integer_
), no_of_games_rescheduled = c(NA, NA, NA, NA, NA, NA), no_of_games_cancelled = c(NA,
NA, NA, NA, NA, NA), mask_mandate = c(NA_integer_, NA_integer_,
NA_integer_, NA_integer_, NA_integer_, NA_integer_), surveillance_testing = c(NA_integer_,
NA_integer_, NA_integer_, NA_integer_, NA_integer_, NA_integer_
), req_daily_health_assessment = c(NA_integer_, NA_integer_,
NA_integer_, NA_integer_, NA_integer_, NA_integer_), precaution_score = c(NA_integer_,
NA_integer_, NA_integer_, NA_integer_, NA_integer_, NA_integer_
)), row.names = c(NA, 6L), class = "data.frame")
CodePudding user response:
There seem to be several things going wrong here that corrplot cannot fix. To note:
- You only have six observations.
- The variables are identical in many cases (how are wins and losses always equal and why are the grouping variables always the same)?
- The data you are trying to pass into a correlation matrix is both categorical and numerical, so it gets confused trying to run it the way you are attempting to.
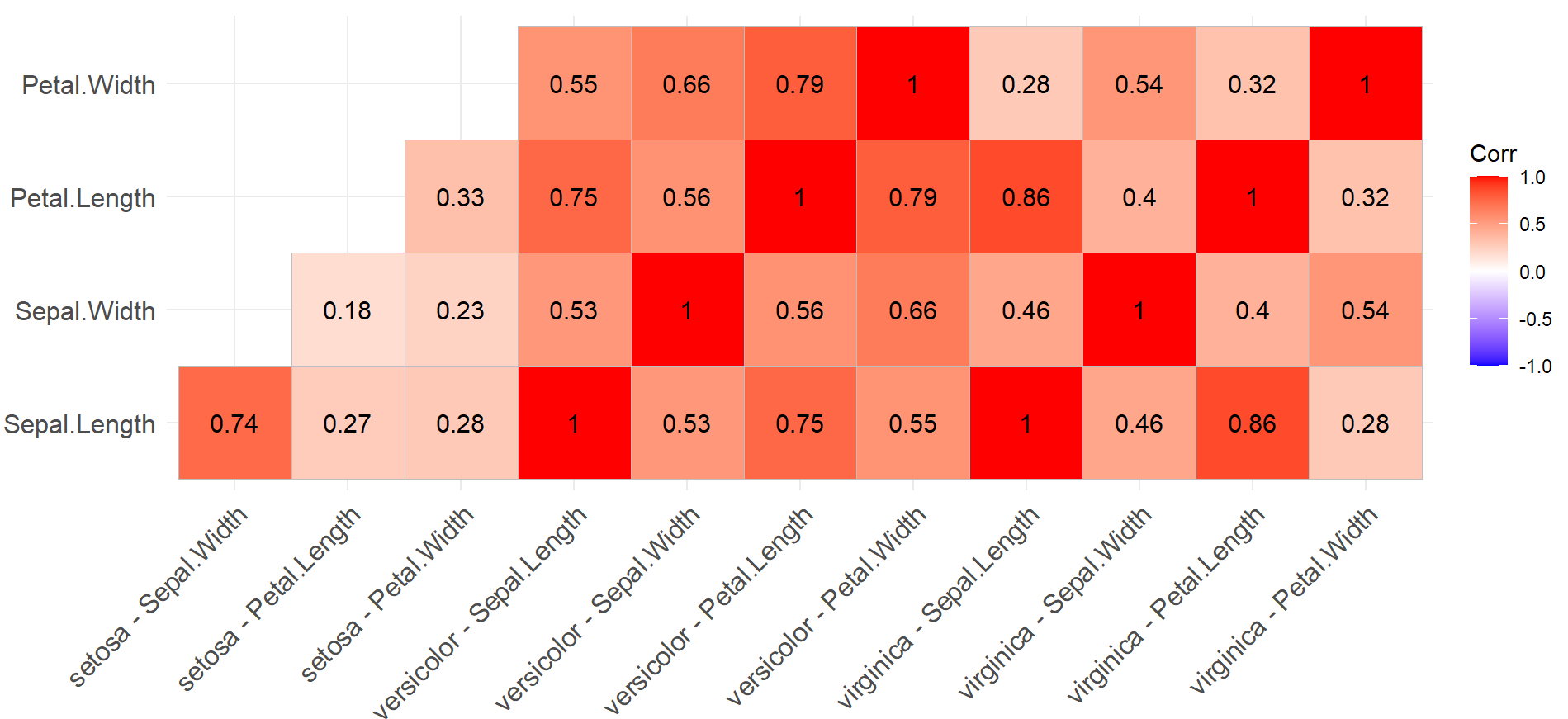
To show you how it works on more typical data, here is how I ran a correlation matrix using the iris data instead:
#### Load Libraries ####
library(tidyverse)
library(ggcorrplot)
library(correlation)
#### Create Correlation Matrix ####
cor.iris <- iris %>%
group_by(Species) %>%
correlation()
#### Run Heatmap ####
ggcorrplot(cor.iris,
type = "lower",
lab = T)
Giving me a grouped correlation with sensible data:
I think the solution here is to find out why your data looks odd, fix it, and then come back to run correlation plots when it looks better.