I made a graph of some GPS-data, where the colour of the path indicates in which habitat a bird was. I added a column to the df, with the desired colours as I wanted to set those manually per habitat type. This is the code I used (which works):
worldmap <- ne_countries(scale = 'medium', type = 'map_units',
returnclass = 'sf')
xlim <- c(min(random_bird$location.long)-0.1, max(random_bird$location.long) 0.1)
ylim <- c(min(random_bird$location.lat)-0.1, max(random_bird$location.lat) 0.1)
ggplot() geom_sf(data = worldmap)
coord_sf(xlim = xlim, ylim = ylim, expand = FALSE)
theme_bw()
geom_path(aes(x = random_bird$location.long, y = random_bird$location.lat), colour = (random_bird$colour))
labs(x="Longitude", y="Latitude") ggtitle(random_bird$colour_ring) theme(plot.title = element_text(hjust = 0.5))
This is a snip of the dataframe:
timestamp location.long location.lat habitat colour colour_ring
2022-08-26 14:34:33.000 2.732021 51.15316 Not classified white S.ROP
2022-08-26 19:34:30.000 2.735457 51.15966 Intertidal cornsilk S.ROP
2022-08-27 03:34:31.000 2.741388 51.16101 Intertidal cornsilk S.ROP
2022-08-27 05:34:30.000 2.736342 51.15954 Intertidal cornsilk S.ROP
2022-08-27 08:34:56.000 2.728476 51.15704 Intertidal cornsilk S.ROP
2022-08-27 10:34:28.000 2.726892 51.15534 Intertidal cornsilk S.ROP
2022-08-27 11:34:28.000 2.726382 51.15453 Not classified white S.ROP
2022-08-27 12:34:27.000 2.726405 51.15470 Not classified white S.ROP
2022-08-27 17:34:32.000 2.723473 51.15557 Intertidal cornsilk S.ROP
2022-08-28 08:34:55.000 2.757212 51.16779 Intertidal cornsilk S.ROP
Now I would like to add a legend mapping the habitat types to the specified colour, I tried using scale_colour_manual(), but can't seem to get it right. Any hints?
CodePudding user response:
If you want a legend then map on aesthetics, i.e. move color=... inside aes. And as your color column already contains the colors use scale_color_identity(guide="legend") to add the colors and a legend. Additionally you have to add nice labels when going this way.
For this reason IMHO it would be easier to map habitat directly on the color aes and then use scale_color_manual to set your desired colors.
Moreover, using random_bird$... is strongly discouraged. Just use column names directly when mapping on aes and add the data for the geom_path via data=random_bird.
Note: I added a grey background to the legend keys to make the colors visible.
library(rnaturalearth)
library(ggplot2)
worldmap <- ne_countries(
scale = "medium", type = "map_units",
returnclass = "sf"
)
xlim <- range(random_bird$location.long) 0.1 * c(-1, 1)
ylim <- range(random_bird$location.lat) 0.1 * c(-1, 1)
base <- ggplot()
geom_sf(data = worldmap)
coord_sf(xlim = xlim, ylim = ylim, expand = FALSE)
theme_bw()
labs(x = "Longitude", y = "Latitude", title = unique(random_bird$colour_ring))
theme(plot.title = element_text(hjust = 0.5), legend.key = element_rect(fill = "grey80"))
base
geom_path(data = random_bird, aes(x = location.long, y = location.lat, colour = colour))
scale_color_identity(guide = "legend")
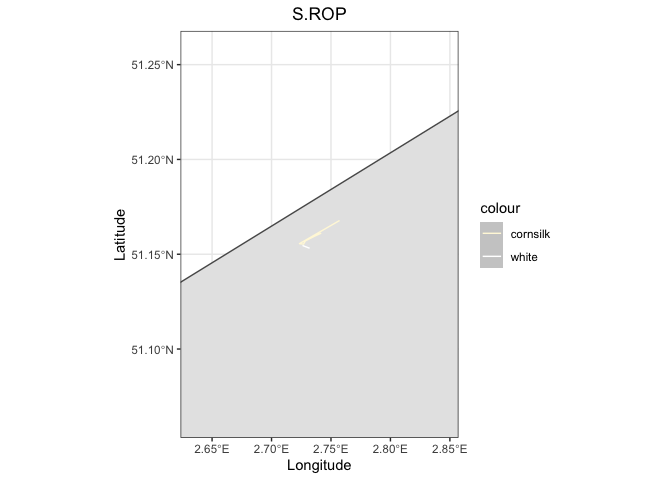
pal <- random_bird |>
dplyr::distinct(habitat, colour) |>
tibble::deframe()
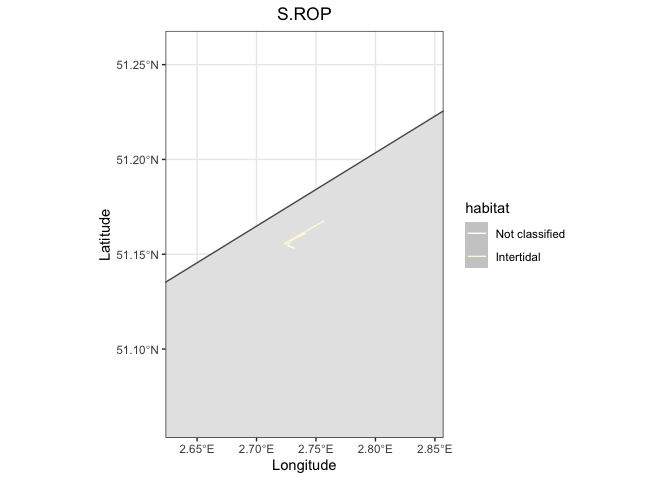
base
geom_path(data = random_bird, aes(x = location.long, y = location.lat, colour = habitat))
scale_color_manual(values = pal)
DATA
random_bird <- structure(list(timestamp = c(
" 2022-08-26 14:34:33.000", " 2022-08-26 19:34:30.000",
"2022-08-27 03:34:31.000", "2022-08-27 05:34:30.000", "2022-08-27 08:34:56.000",
"2022-08-27 10:34:28.000", "2022-08-27 11:34:28.000", "2022-08-27 12:34:27.000",
"2022-08-27 17:34:32.000", "2022-08-28 08:34:55.000"
), location.long = c(
2.732021,
2.735457, 2.741388, 2.736342, 2.728476, 2.726892, 2.726382, 2.726405,
2.723473, 2.757212
), location.lat = c(
51.15316, 51.15966, 51.16101,
51.15954, 51.15704, 51.15534, 51.15453, 51.1547, 51.15557, 51.16779
), habitat = c(
"Not classified", "Intertidal", "Intertidal",
"Intertidal", "Intertidal", "Intertidal", "Not classified", "Not classified",
"Intertidal", "Intertidal"
), colour = c(
"white", "cornsilk",
"cornsilk", "cornsilk", "cornsilk", "cornsilk", "white", "white",
"cornsilk", "cornsilk"
), colour_ring = c(
"S.ROP", "S.ROP", "S.ROP",
"S.ROP", "S.ROP", "S.ROP", "S.ROP", "S.ROP", "S.ROP", "S.ROP"
)), class = "data.frame", row.names = c(NA, -10L))
