I have the names of the files I want to concatenate: files<-c("test/df1.tsv" "test/df2.tsv")
I have x2 .tsv files that look like so (sorry - I didn't know how to code this into an reproducible, example df):
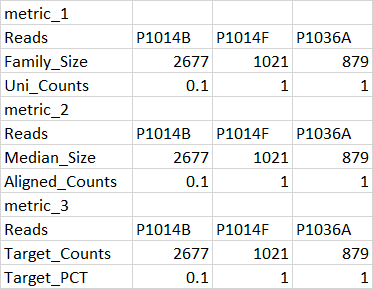
df1
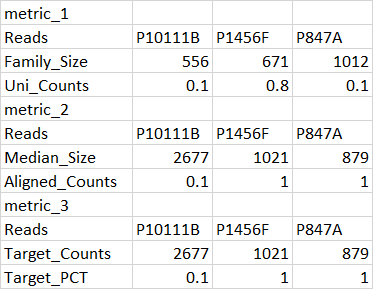
df2
I am trying to concatenate these tsv files by column. I have managed to read some blogs and have got so far as using map:
metrics_df <- map_df(files, read_tsv)
But this is doing an rbind like so:
metrics_df
# A tibble: 22 × 4
metric_1 ...2 ...3 ...4
<chr> <chr> <chr> <chr>
1 Reads P1014B P1014F P1036A
2 Family_Size 2677 1021 879
3 Uni_Counts 0.1 1 1
4 metric_2 NA NA NA
5 Reads P1014B P1014F P1036A
6 Median_Size 2677 1021 879
7 Aligned_Counts 0.1 1 1
8 metric_3 NA NA NA
9 Reads P1014B P1014F P1036A
10 Target_Counts 2677 1021 879
11 Target_PCT 0.1 1 1
12 Reads P10111B P1456F P847A
13 Family_Size 556 671 1012
14 Uni_Counts 0.1 0.8 0.1
15 metric_2 NA NA NA
16 Reads P10111B P1456F P847A
17 Median_Size 2677 1021 879
18 Aligned_Counts 0.1 1 1
19 metric_3 NA NA NA
20 Reads P10111B P1456F P847A
21 Target_Counts 2677 1021 879
22 Target_PCT 0.1 1 1
I would like to combine the files so that the columns are the concatenated 'reads' row.
Is there a way of doing this with map() or alternatively splitting the df using dplyr???
CodePudding user response:
You can try this:
df <- list.files(path='$YOUR_FILES_PATH') %>%
lapply(read_tsv) %>%
bind_rows
CodePudding user response:
A simple way would be:
Lines = c()
for(f in files) { Lines = c(Lines, readLines(f)) }
writeLines(Lines, "CombinedFiles.tsv")