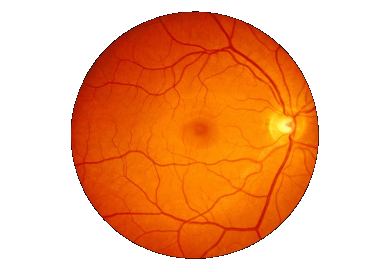
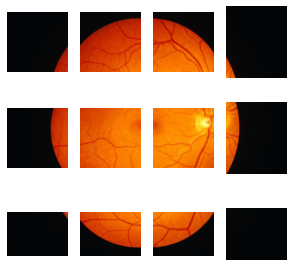
Say I have an image as follows and I would like to crop some sub-images of the retina only.
So the first step I took is to find a mask
# Find Mask
img = imread(folder 'test.jpg')
blur = cv2.GaussianBlur(img, (3, 3), cv2.BORDER_DEFAULT)
edged = cv2.Canny(blur, 10, 250)
# threshold
thresh = cv2.threshold(edged, 128, 255, cv2.THRESH_BINARY)[1]
# apply close morphology
kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (5,5))
thresh = cv2.morphologyEx(thresh, cv2.MORPH_CLOSE, kernel)
# get bounding box coordinates from the one filled external contour
filled = np.zeros_like(thresh)
cnts = cv2.findContours(thresh, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
cnts = cnts[0] if len(cnts) == 2 else cnts[1]
max_cnt = max(cnts, key=cv2.contourArea)
x,y,w,h = cv2.boundingRect(max_cnt)
cv2.drawContours(filled, [max_cnt], 0, 255, -1)
# crop filled contour image
mask = filled.copy()
cv2_imshow(mask)
Then I wish to crop subimages of 50 * 50 within the inner area of a mask
Here is some code I found online:
# This gives the retina only
new_image = cv2.cvtColor(img, cv2.COLOR_BGR2BGRA)
new_image[:,:,3] = mask[:,:]
imshow(new_image)
Below is closer to what I want. There are two sub-images that are within the mask. But what I really want is to crop maybe 5 sub-images randomly that are within the mask. Is there a way that I can do that? Or how can I check the cropped images are within the mask?
def img_to_grid(img):
# ww = [[i.min(), i.max()] for i in np.array_split(range(img.shape[0]),row)]
# hh = [[i.min(), i.max()] for i in np.array_split(range(img.shape[1]),col)]
print(img.shape[0], img.shape[1])
ww = [[i, i 100-1] for i in range(0, img.shape[0],100)]
print(ww)
hh = [[i, i 100-1] for i in range(0, img.shape[1],100)]
print(hh)
grid = [img[j:jj,i:ii,:] for j,jj in ww for i,ii in hh]
print((j, jj, i, ii) for j,jj in ww for i,ii in hh)
return grid, len(ww), len(hh)
def plot_grid(grid,row,col,h=5,w=5):
fig, ax = plt.subplots(nrows=row, ncols=col)
[axi.set_axis_off() for axi in ax.ravel()]
fig.set_figheight(h)
fig.set_figwidth(w)
c = 0
for row in ax:
for col in row:
col.imshow(np.flip(grid[c],axis=-1))
c =1
plt.show()
grid , r,c = img_to_grid(img)
plot_grid(grid,r,c)
CodePudding user response:
If you want to extract really random sub-images that is not necessary inscribed in a circle, you can just get some coords within the non-zero mask bounding box, check if it contains only ones, and save them, if so.
sub_imgs = []
# Define minimum width and height for the cropped region
min_width = 32
min_height = 32
# Define the number of sub-images
sub_img_count = 5
while len(sub_imgs) < sub_img_count:
x1 = np.random.randint(x, x w)
y1 = np.random.randint(y, y h)
maxw = w - (x1 - x)
maxh = h - (y1 - y)
# Skip if maximum possible sizes don't meet the requirement
if not (maxw > min_width and maxh > min_height):
continue
width = np.random.randint(min_width, maxw)
height = np.random.randint(min_height, maxh)
# Extract the region only if it is within the mask
if np.all(mask[y1: y1 height, x1: x1 width]):
sub_img = img[y1: y1 height, x1: x1 width, :]
sub_imgs.append(sub_img)
# Save the cropped regions
for i, sub_img in enumerate(sub_imgs):
cv2.imwrite(f'{i 1}.jpg', sub_img)