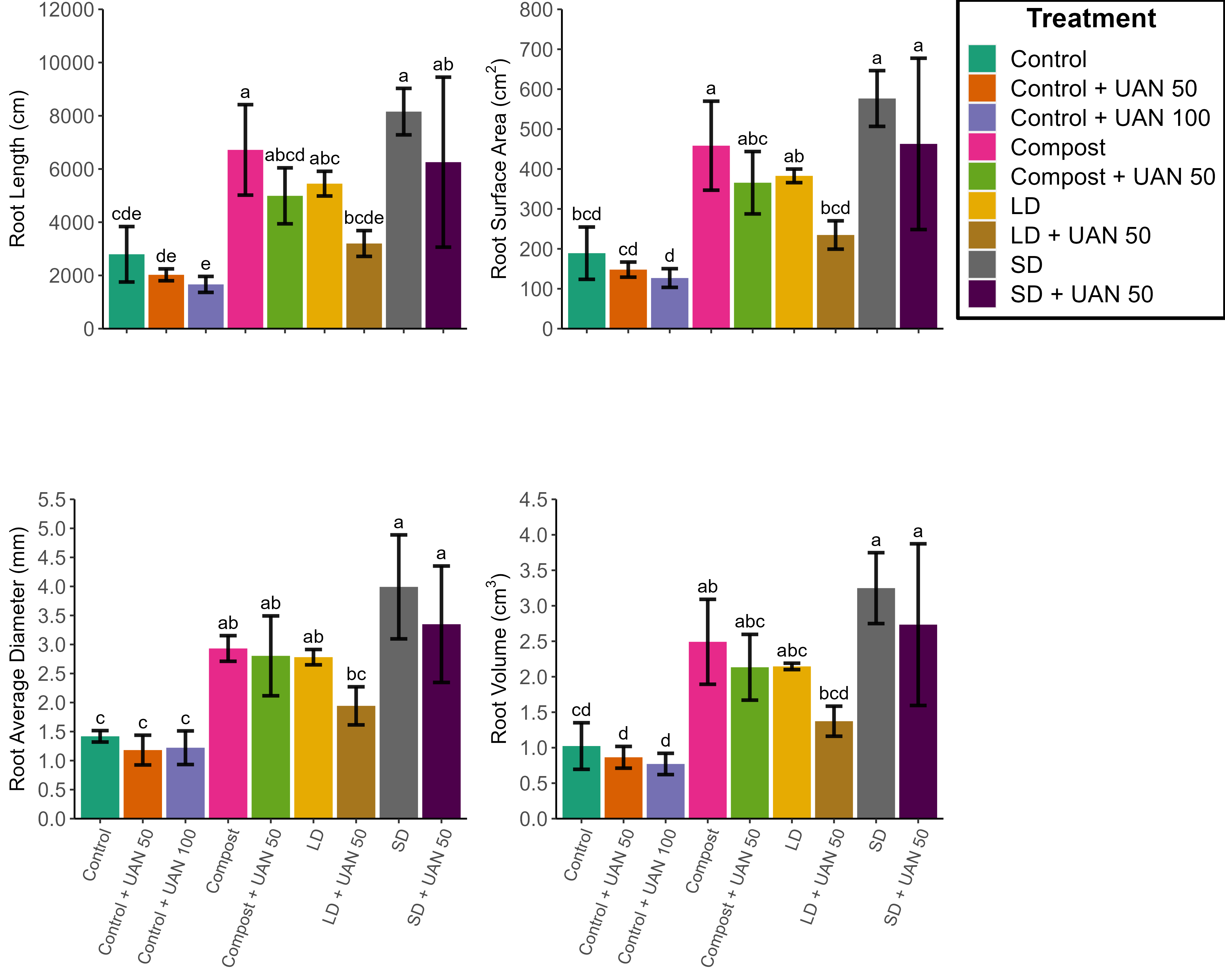
As the title suggests I am trying to remove the gap where the x axis labels are meant to be for the top graphs - Root length and Root Surface Area. Any suggestions?
CODE
#Tukeys lettering
anova <- aov(Root_Length ~Treatment, data =JO_Data)
tukey <- TukeyHSD(anova)
cld <- multcompLetters4(anova, tukey)
JO_Data_Root_Length <- JO_Data %>% dplyr::group_by(Treatment) %>% dplyr::summarise(mean=mean(Root_Length), sd = sd(Root_Length)) %>% arrange(desc(mean))
cld <- as.data.frame.list(cld$Treatment)
JO_Data_Root_Length$cld <- cld$Letters
# Root length plot
plot.JO_Data.Root_Length=ggplot(JO_Data_Root_Length,aes(x = Treatment, y = mean, fill=Treatment))
geom_bar(aes(x=Treatment, y=mean), stat="identity", alpha=1)
geom_errorbar( aes(x = Treatment, ymin = mean -sd, ymax = mean sd), width=0.4, colour="black", alpha=0.9, size=1)
geom_text(aes(label = cld, y = mean sd), vjust = -0.5)
labs(y = expression(paste("Root Length (cm)")))
scale_y_continuous(expand = c(0,0), limits = c(0,12000), breaks = seq(0, 12000, by = 2000))
scale_x_discrete(limits = c("Control", "Control UAN 50", "Control UAN 100","Compost", "Compost UAN 50", "LD", "LD UAN 50", "SD", "SD UAN 50"))
scale_fill_manual(values = cbbPalette, breaks = c("Control", "Control UAN 50", "Control UAN 100","Compost", "Compost UAN 50", "LD", "LD UAN 50", "SD", "SD UAN 50"))
theme(axis.line = element_line(colour = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_text(size = 12),
axis.title.y = element_text(size = 12))
theme(legend.justification = c("right", "top"))
theme(legend.title = element_text(color = "black", size = 16, face = "bold"))
theme(legend.title.align = 0.5)
theme(legend.background = element_rect(linetype = "solid", colour = "black", size = 1))
theme(legend.margin = margin(0.1,0.2,0.2,0.2, unit ="cm"))
theme(legend.text = element_text(size=14, colour="black"))
theme(axis.title.x=element_blank())
plot.JO_Data.Root_Length
################### three other graphs not added in code to reduce size but is the same as Root length for the rest of the graphs ########
# Putting all the plots together
# install.packages("gridExtra")
library(gridExtra)
# install.packages("cowplot")
library(cowplot)
?plot_grid
JO_plot2 <- plot_grid(plot.JO_Data.Root_Length theme(legend.position="none"),
plot.JO_Data.Root_SA theme(legend.position="none"),
plot.JO_Data.Root_diameter theme(legend.position="none"),
plot.JO_Data.Root_Vol theme(legend.position="none"),
align = "h",
axis = "b",
hjust = 0.1,
vjust = 0.1,
nrow = 2,
ncol =2)
legend <- get_legend(plot.JO_Data.Root_Length)
Root_plot2 <- plot_grid(JO_plot2, legend, rel_widths = c(1.75, 0.5))
Root_plot2
Graph Output
I thought the axis.text.x = element_blank() would fix the problem but it hasn't.
Cheers :)
CodePudding user response:
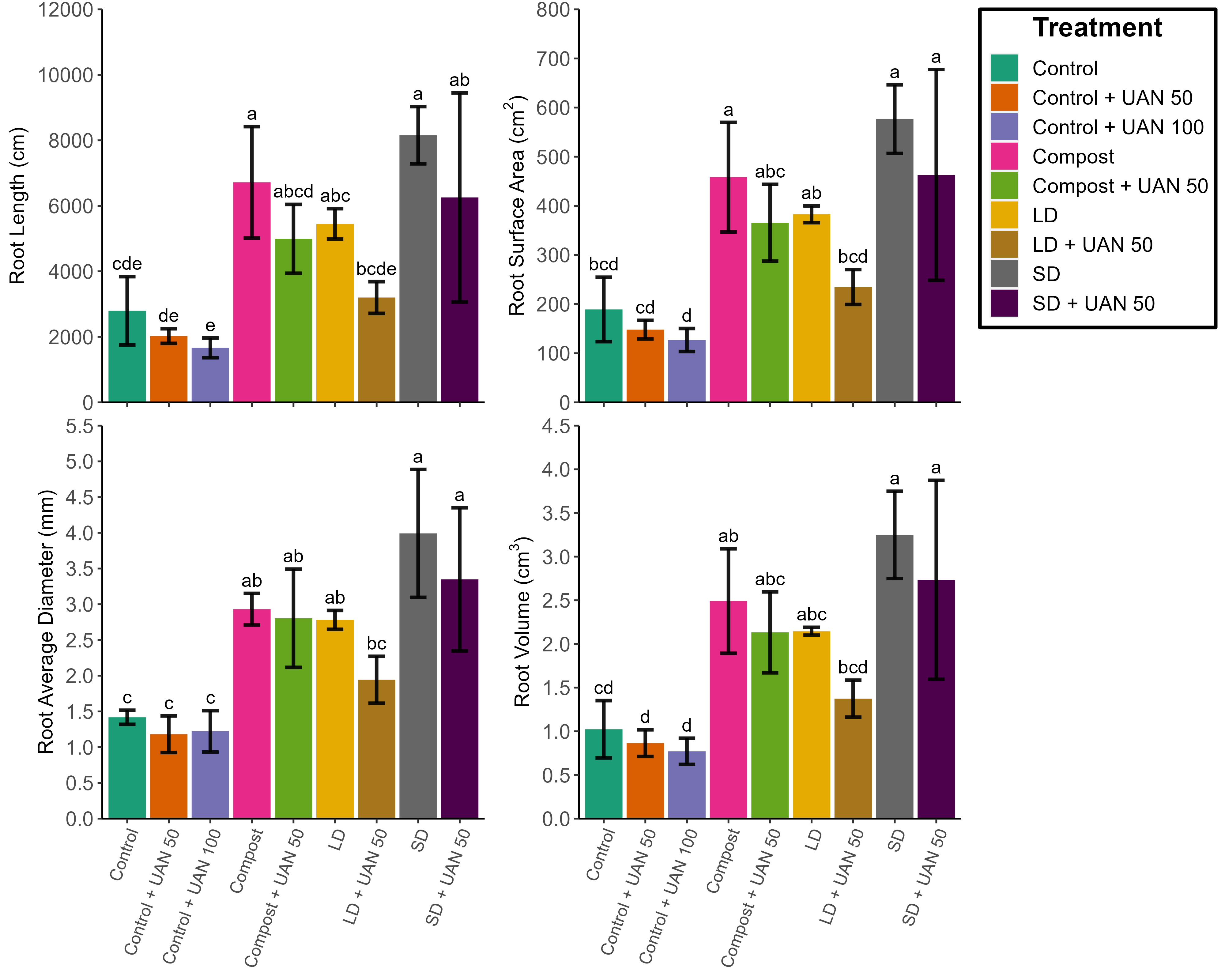
Instead of plot_grid, I used:
ggarrange(plot.JO_Data.Root_Length theme(legend.position="none"),
plot.JO_Data.Root_SA,
plot.JO_Data.Root_diameter theme(legend.position="none"),
plot.JO_Data.Root_Vol theme(legend.position="none"), ncol = 2)
Image
seemed to fix this, however, would be good to use a command to remove that gap in plot_grid
CodePudding user response:
You could provide plot.margin for both plots and set negative value for the bottom margin and upper margin. This will ensure that both plot joins.