I have the following dataset (the outpur is generated with dput):
structure(list(Var1 = structure(c(1L, 1L, 2L, 1L, 2L, 3L, 1L,
2L, 3L, 4L, 1L, 2L, 3L, 4L, 5L, 1L, 2L, 3L, 4L, 5L, 6L, 1L, 2L,
3L, 4L, 5L, 6L, 7L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 1L, 2L, 3L,
4L, 5L, 6L, 7L, 8L, 9L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 1L, 2L, 3L, 4L,
5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L,
8L, 9L, 10L, 11L, 12L, 13L), levels = c("X0", "X1", "X2", "X3",
"X4", "X5", "X6", "X7", "X8", "maxd", "lag", "tas", "mean"), class = "factor"),
Var2 = c("X0", "X1", "X1", "X2", "X2", "X2", "X3", "X3",
"X3", "X3", "X4", "X4", "X4", "X4", "X4", "X5", "X5", "X5",
"X5", "X5", "X5", "X6", "X6", "X6", "X6", "X6", "X6", "X6",
"X7", "X7", "X7", "X7", "X7", "X7", "X7", "X7", "X8", "X8",
"X8", "X8", "X8", "X8", "X8", "X8", "X8", "maxd", "maxd",
"maxd", "maxd", "maxd", "maxd", "maxd", "maxd", "maxd", "maxd",
"lag", "lag", "lag", "lag", "lag", "lag", "lag", "lag", "lag",
"lag", "lag", "tas", "tas", "tas", "tas", "tas", "tas", "tas",
"tas", "tas", "tas", "tas", "tas", "mean", "mean", "mean",
"mean", "mean", "mean", "mean", "mean", "mean", "mean", "mean",
"mean", "mean"), value = c(1, 0, 1, 0, 0, 1, 0, 0, 0, 1,
0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0,
0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, -0.369, -0.068,
-0.263, -0.173, 0.042, -0.146, 0.077, 0.053, 0.09, 1, -0.15,
0.021, -0.124, -0.086, -0.011, 0.008, 0.043, 0.005, 0.068,
0.677, 1, -0.03, -0.149, 0.04, 0.019, 0.078, 0.06, 0.003,
-0.022, 0.089, 0.283, 0.247, 1, -0.101, -0.274, -0.176, -0.033,
0.08, 0.009, 0.081, 0.062, 0.154, 0.509, 0.49, 0.394, 1),
vif = c(1.60070614254443, 1.11225298408558, 1.11225298408558,
1.24667893632764, 1.24667893632764, 1.24667893632764, 1.13718634714263,
1.13718634714263, 1.13718634714263, 1.13718634714263, 1.06477651305619,
1.06477651305619, 1.06477651305619, 1.06477651305619, 1.06477651305619,
1.15665232275605, 1.15665232275605, 1.15665232275605, 1.15665232275605,
1.15665232275605, 1.15665232275605, 1.04455882370308, 1.04455882370308,
1.04455882370308, 1.04455882370308, 1.04455882370308, 1.04455882370308,
1.04455882370308, 1.0752045744496, 1.0752045744496, 1.0752045744496,
1.0752045744496, 1.0752045744496, 1.0752045744496, 1.0752045744496,
1.0752045744496, 1.07921364011364, 1.07921364011364, 1.07921364011364,
1.07921364011364, 1.07921364011364, 1.07921364011364, 1.07921364011364,
1.07921364011364, 1.07921364011364, 2.8566720117982, 2.8566720117982,
2.8566720117982, 2.8566720117982, 2.8566720117982, 2.8566720117982,
2.8566720117982, 2.8566720117982, 2.8566720117982, 2.8566720117982,
2.09930904742747, 2.09930904742747, 2.09930904742747, 2.09930904742747,
2.09930904742747, 2.09930904742747, 2.09930904742747, 2.09930904742747,
2.09930904742747, 2.09930904742747, 2.09930904742747, 1.43215872939721,
1.43215872939721, 1.43215872939721, 1.43215872939721, 1.43215872939721,
1.43215872939721, 1.43215872939721, 1.43215872939721, 1.43215872939721,
1.43215872939721, 1.43215872939721, 1.43215872939721, 2.21146503479172,
2.21146503479172, 2.21146503479172, 2.21146503479172, 2.21146503479172,
2.21146503479172, 2.21146503479172, 2.21146503479172, 2.21146503479172,
2.21146503479172, 2.21146503479172, 2.21146503479172, 2.21146503479172
)), class = "data.frame", row.names = c(NA, -91L))
and I have created the heatmap of the correlations:
ggplot(data = melted_cormat, aes(Var2, Var1, fill = value))
geom_tile(color = "white")
scale_fill_gradient2(low = "#2bc290", high = "#b6c1ff", mid = "#F9F7F5",
midpoint = 0, limit = c(-1,1), space = "Lab",
name="Magnitude of\nCorrelation")
theme_minimal()
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 12, hjust = 1))
coord_fixed() xlab("") ylab("")
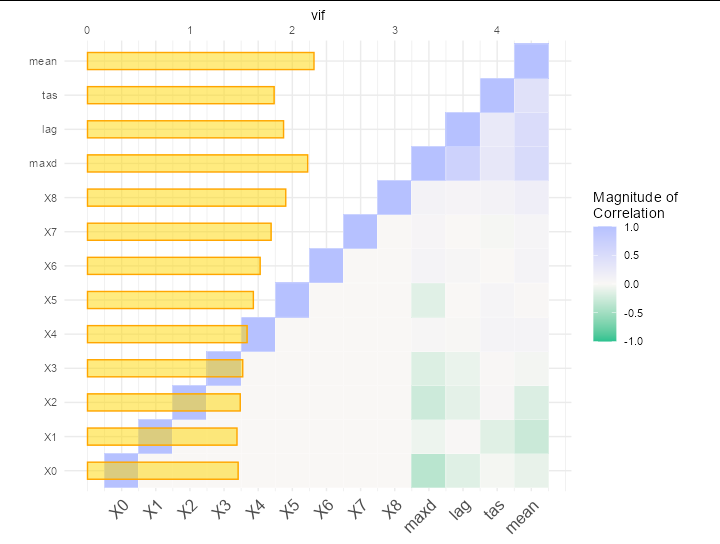
But I would like to add a barplot to the plot in order to look like that:
Can I do that?
CodePudding user response:
This is possible (though not advisable since it makes for a messy and confusing plot). The secret is to make the x axis numeric, but give it the factor levels of Var2 as labels. Then plot vif as a bar plot and use a secondary axis to label it.
Start by explicitly converting Var2 to a factor:
melted_cormat$Var2 <- factor(melted_cormat$Var2,
levels = c(paste0('X', 0:8), 'maxd', 'lag',
'tas', 'mean'))
And the plotting code is like this:
ggplot(data = melted_cormat, aes(as.numeric(Var2), Var1, fill = value))
geom_tile(color = "white")
geom_bar(stat = 'summary', fun = mean,
aes(y = Var1, x = vif * 3), fill ='gold', position = 'identity',
width = 0.5, color = 'orange', alpha = 0.5)
scale_fill_gradient2(low = "#2bc290", high = "#b6c1ff", mid = "#F9F7F5",
midpoint = 0, limit = c(-1,1), space = "Lab",
name="Magnitude of\nCorrelation")
scale_x_continuous(breaks = seq_along(levels(melted_cormat$Var2)),
labels = levels(melted_cormat$Var2),
sec.axis = sec_axis(~.x/3, name = 'vif'))
theme_minimal()
theme(axis.text.x.bottom = element_text(angle = 45, vjust = 1,
size = 12, hjust = 1))
coord_fixed()
labs(y = NULL, x = NULL)