I'm new to R studio and I need to plot a directed network. I have a node and edge .csv files
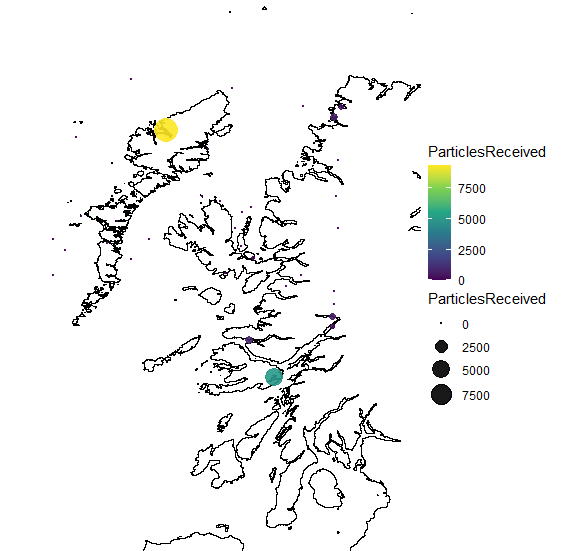
So far I've just been able to plot in the map the points receiving particles
I'm facing several issues here. The first one is that not all the points receiving particles are properly observed in the map because of the color code and dimension of the circles.
The second issue is that I also need to plot the target points, but I only manage to plot those receiving particles.
And I need to connect source points with target points (connected by lines more or less thickness depending on the particles sended by the source point).
This is my code:
require(rgdal)
require(ggplot2)
fn <- file.path(tempdir(), "GBR_adm_gdb.zip", fsep = "\\")
download.file("http://biogeo.ucdavis.edu/data/gadm2.8/shp/GBR_adm_shp.zip", fn)
utils::unzip(fn, exdir = tempdir())
shp <- readOGR(dsn = file.path(tempdir(), "GBR_adm1.shp"), stringsAsFactors = F)
nodes <- read.csv("C:/Users/.../NODES.csv" , header=TRUE)
head(nodes)
edges <- read.csv("C:/Users/.../edges.csv" , header=TRUE)
head(edges)
map <- ggplot() geom_polygon(data = shp, aes(x = long, y = lat, group = group), colour = "black", fill = NA)
geom_point (data = nodes,
aes(x = as.numeric(Longitude),
y = as.numeric(Latitude), size = ParticlesReceived, color =ParticlesReceived), alpha = .9)
geom_line()
scale_size_area(max_size =8)
scale_color_viridis_c()
coord_fixed(ratio = 1.3,
xlim = c(-8,-4.5),
ylim = c(55.3, 59))
theme(legend.position = 'none')
Thank you in advance for your help.
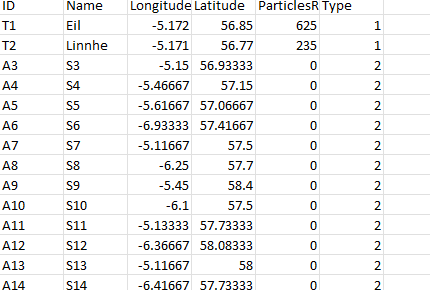
nodes <- structure(list(ID = c("T1", "T2", "A3", "A4", "A5", "A6", "A7",
"A8", "A9", "A10", "A11", "A12", "A13", "A14", "A15", "A16",
"A17_P", "A18", "A19", "A20", "A21", "A22", "A23", "A24", "A25",
"A26", "A27", "A28", "A29", "A30", "T3", "T7", "T4", "T5", "T6",
"T8", "A31", "A32", "A33", "A34", "A35", "A36", "A37", "A38",
"A39", "A40", "A41", "A42", "A43", "A44", "A45", "A46", "A47",
"A48", "A49", "A50", "A51", "A52", "A53", "A54", "A55", "A56",
"A57", "A58", "A59", "A60", "A61", "A62", "A63_B", "", "", ""
), Name = c("Eil", "Linnhe", "S3", "S4", "S5", "S6", "S7", "S8",
"S9", "S10", "S11", "S12", "S13", "S14", "S15", "S16", "S17_P",
"S18", "S19", "S20", "S21", "S22", "S23", "S24", "S25", "S26",
"S27", "S28", "S29", "S30", "Sunart", "Spelve", "Roag", "Badcall",
"Laxford", "Cairidih", "S31", "S32", "S33", "S34", "S35", "S36",
"S37", "S38", "S39", "S40", "S41", "S42", "S43", "S44", "S45",
"S46", "S47", "S48", "S49", "S50", "S51", "S52", "S53", "S54",
"S55", "S56", "S57", "S58", "S59", "S60", "S61", "S62", "S63_B",
"", "", ""), Longitude = c(-5.172, -5.171, -5.973, -5.279, -5.371,
-6.557, -5.79, -6.612, -7.511, -7.203, -7.137, -7.35, -5.909,
-6.016, -6.028, -6.119, -6.147, -5.266, -6.215, -5.882, -5.743,
-5.845, -5.467, -7.639, -7.596, -7.063, -7.507, -7.375, -7.436,
-6.817, -5.728755, -5.965663, -6.768254, -5.151579, -5.082169,
-5.931472, -6.34, -6.892, -5.946, -6.255, -6.324, -5.515, -6.458,
-6.284, -6.427, -6.399, -5.631, -6.768, -6.04, -6.274, -5.946,
-6.435, -7.35, -5.968, -6.573, -6.762, -6.29, -7.13, -7, -7.764,
-6.935, -6.966, -5.663, -6.964, -7.315, -5.973, -6.685, -6.559,
-7.391053, NA, NA, NA), Latitude = c(56.85, 56.77, 56.56, 56.69,
56.64, 57.25, 57.3, 57.3, 56.85, 57.23, 57.33, 56.95, 57.02,
56.97, 56.82, 57.1, 57.42, 58.24, 57.65, 57.44, 57.6, 57.44,
57.39, 56.75, 56.76, 57.96, 57.09, 57.7, 57.2, 58.32, 56.39908,
56.668643, 58.220343, 58.316558, 58.394389, 57.277806, 56.24,
56.44, 56.05, 56.54, 56.55, 56.55, 56.61, 56.2, 56.18, 56.27,
56.47, 56.5, 55.98, 56.24, 56.05, 56.36, 56.95, 56.9, 57.03,
57.39, 57.13, 56.37, 56.17, 56.38, 55.93, 55.78, 56.05, 56.42,
56.53, 56.56, 56.44, 56.58, 56.997312, NA, NA, NA), ParticlesReceived = c(625L,
235L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
326L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 4951L,
829L, 9291L, 774L, 454L, 145L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 2768L, NA, NA, NA), Type = c(1L,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, NA, NA, NA)), row.names = c(NA, 72L), class = "data.frame")
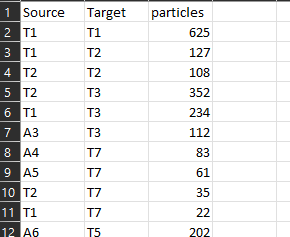
edges <- structure(list(Source = c("T1", "T1", "T2", "T2", "T1", "A3",
"A4", "A5", "T2", "T1", "A6", "A7", "A17_P", "A18", "A8", "A20",
"A19", "A8", "A18", "A20", "A6", "A21", "A7", "A22", "A25", "A9",
"A10", "A26", "A27", "A11", "A12", "A24", "A28", "A29", "A30",
"A15", "A13", "A14", "A16", "A23", "T3", "T7", "T4", "T8", "T6",
"A31", "A32", "A33", "A34", "A35", "A36", "A37", "A38", "A39",
"A40", "A41", "A42", "A43", "A44", "A45", "A46", "A47", "A48",
"A49", "A50", "A51", "A52", "A53", "A54", "A55", "A56", "A57",
"A58", "A59", "A60", "A61", "A62"), Target = c("T1", "T2", "T2",
"T3", "T3", "T3", "T7", "T7", "T7", "T7", "T5", "T5", "T5", "T5",
"T5", "T5", "T5", "T6", "T6", "T6", "T6", "T6", "T6", "T6", "T4",
"T4", "T4", "T4", "T4", "T4", "T4", "T4", "T4", "T4", "T4", "T8",
"T8", "T8", "T8", "T8", "T3", "T7", "T4", "T5", "T6", "A63_B",
"A63_B", "A63_B", "A63_B", "A63_B", "A63_B", "A63_B", "A63_B",
"A63_B", "A63_B", "A63_B", "A63_B", "A63_B", "A63_B", "A63_B",
"A63_B", "A63_B", "A63_B", "A63_B", "A63_B", "A63_B", "A63_B",
"A63_B", "A63_B", "A63_B", "A63_B", "A63_B", "A17_P", "A17_P",
"A17_P", "A17_P", "A17_P"), particles = c(625L, 127L, 108L, 352L,
234L, 112L, 83L, 61L, 35L, 22L, 202L, 186L, 154L, 77L, 75L, 41L,
32L, 160L, 47L, 46L, 40L, 40L, 23L, 19L, 504L, 336L, 299L, 294L,
275L, 168L, 168L, 168L, 168L, 152L, 136L, 52L, 43L, 20L, 19L,
11L, 4365L, 628L, 6623L, 7L, 79L, 143L, 67L, 37L, 29L, 300L,
87L, 142L, 194L, 38L, 56L, 108L, 33L, 25L, 148L, 101L, 51L, 211L,
152L, 336L, 39L, 24L, 109L, 84L, 90L, 36L, 35L, 93L, 33L, 67L,
134L, 60L, 32L)), row.names = c(NA, 77L), class = "data.frame")
CodePudding user response:
To draw edges between node you're better off transforming your data to a graph.
require(tidygraph)
particles_graph <- tbl_graph(nodes, edges, node_key= "ID")
Once you have it in a graph format, you can use the ggraph package.
We first have to create a manual layout with the coordinate system:
particle_coord_layout <- create_layout(graph = particles_graph,
layout = "manual",
x = nodes$Longitude,
y = nodes$Latitude)
And then we can create a crude graph:
ggraph(particle_coord_layout)
geom_polygon(data = shp, aes(x = long, y = lat, group = group), colour = "black", fill = NA)
geom_edge_link(aes(width=particles), alpha = .5)
geom_node_point(aes(size = ParticlesReceived))
coord_fixed(ratio = 1.3,
xlim = c(-7.6,-4.8),
ylim = c(55.8, 58.6))
theme(legend.position = "none")
#> Regions defined for each Polygons
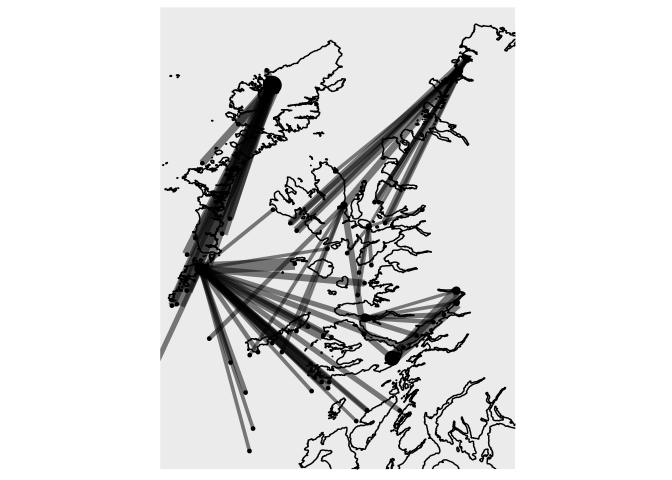
Exploring the ggraph package we can improve the results:
ggraph(particle_coord_layout)
geom_polygon(data = shp, aes(x = long, y = lat, group = group), fill = "grey85")
geom_edge_diagonal(aes(width=particles, alpha=after_stat(index), color=particles))
geom_node_text(aes(label=Name, size = ParticlesReceived), alpha=.8)
scale_edge_width_continuous(range=c(1,3))
scale_edge_color_gradientn(colors = scales::brewer_pal("seq", direction = 1)(5)[3:5])
scale_edge_alpha(range=c(0,1))
scale_size(range=c(2,6))
coord_fixed(ratio = 1.3,
xlim = c(-7.6,-4.8),
ylim = c(55.8, 58.6))
theme_void()
theme(legend.position = "none")
#> Regions defined for each Polygons
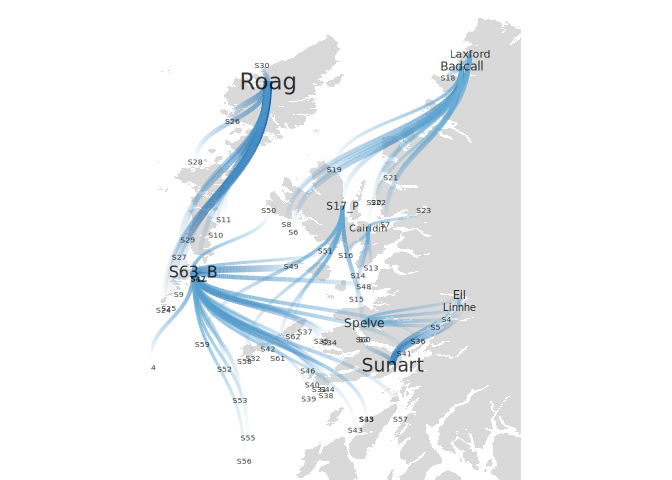
Created on 2022-11-03 with reprex v2.0.2