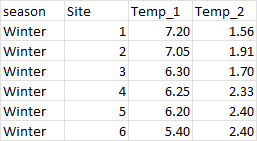
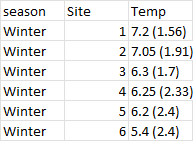
I Have a data frame of several water quality measures. For each measure I have a calculated mean and SD. I have a value for 6 sites and 4 seasons. Currently my dataframe has the means in a column for examples 'Temp_1' and then a column for the standard deviation as 'Temp_2'. I want to export the file with one column for each water quality measure with the format mean (SD).
current output
This is an example for the first water measure, but I'd like to code it so it is also done to remaining factors as well.
desired output
Head of dataframe
structure(list(season = structure(c(1L, 1L, 1L, 1L, 1L, 1L), levels = c("Winter",
"Spring", "Summer", "Autumn"), class = "factor"), Site = structure(1:6, levels = c("1",
"2", "3", "4", "5", "6"), class = "factor"), Temp_1 = c(7.2,
7.05, 6.3, 6.25, 6.2, 5.4), Temp_2 = c(1.55563491861041, 1.90918830920368,
1.69705627484771, 2.33345237791561, 2.40416305603426, 2.40416305603426
), pH_1 = c(7.435, 7.38, 7.52, 7.525, 7.38, 7.565), pH_2 = c(0.289913780286484,
0.282842712474619, 0.0989949493661164, 0.120208152801713, 0.0565685424949239,
0.261629509039023), DO_1 = c(9, 9.1, 8.25, 8.85, 9.25, 9), DO_2 = c(0,
0.424264068711928, 0.0707106781186558, 0.494974746830583, 0.636396103067892,
0.42426406871193), EC_1 = c(337.5, 333, 321.5, 322, 309, 300.5
), EC_2 = c(55.8614357137373, 41.0121933088198, 51.618795026618,
32.5269119345812, 25.4558441227157, 30.4055915910215), SS_1 = c(5.945,
3.65, 5.025, 2.535, 10.22, 4.595), SS_2 = c(0.728319984622144,
1.06066017177982, 2.93449314192417, 0.473761543394987, 8.23072293301141,
0.67175144212722), TP_1 = c(73.5, 75, 61.5, 66.5, 83, 87), TP_2 = c(3.53553390593274,
12.7279220613579, 9.19238815542512, 6.36396103067893, 26.8700576850888,
24.0416305603426), SRP_1 = c(19, 19, 10, 14, 13.5, 23.5), SRP_2 = c(2.82842712474619,
1.4142135623731, 2.82842712474619, 0, 0.707106781186548, 3.53553390593274
), PP_1 = c(54.5, 56, 51.5, 52.5, 69.5, 63.5), PP_2 = c(6.36396103067893,
11.3137084989848, 6.36396103067893, 6.36396103067893, 26.1629509039023,
20.5060966544099), DA_1 = c(0.083, 0.0775, 0.0775, 0.044, 0.059,
0.051), DA_2 = c(0.00282842712474619, 0.0120208152801713, 0.00919238815542513,
0.0014142135623731, 0.0127279220613579, 0.00848528137423857),
DNI_1 = c(0.048739437, 0.041015562, 0.0617723365, 0.0337441755,
0.041480944, 0.0143461675), DNI_2 = c(0.0345079125942686,
0.0223312453226695, 0.0187360224120165, 0.0162032493604065,
0.0258169069873252, 0.0202885446465761), DNA_1 = c(20.43507986,
20.438919615, 14.98692746, 19.953408625, 17.03060377, 8.5767502525
), DNA_2 = c(1.80288106961836, 1.2687128010491, 2.28839365291436,
1.03116172040732, 0.396528484042397, 1.72350828181138), DF_1 = c(0.0992379715,
0.0947268395, 0.094323125, 0.098064875, 0.0980304675, 0.085783911
), DF_2 = c(0.00372072305060515, 0.00724914346231915, 0.0142932471712976,
0.0116895470668939, 0.00255671780854136, 0.00830519117656529
), DC_1 = c(12.18685357, 12.73924378, 13.09550326, 13.417557825,
15.140975265, 21.429763715), DC_2 = c(0.57615880774946, 0.0430071960969884,
0.702539578486863, 0.134642528587041, 0.66786605299916, 0.17012889453292
), DS_1 = c(15.834380095, 15.69623116, 14.37636388, 15.444235935,
14.647596185, 11.9877372), DS_2 = c(1.67153135346354, 1.69978765863781,
2.47560570280853, 1.03831263471691, 1.24488755930594, 0.975483163720397
), DOC_1 = c(19.74, 20.08, 21.24, 20.34, 21.88, 24.92), DOC_2 = c(2.7435743110038,
1.69705627484772, 2.60215295476649, 1.04651803615609, 0.226274169979695,
0.452548339959388)), row.names = c(NA, 6L), class = "data.frame")
CodePudding user response:
Using mutate across with some tricks to organize paired data we can do it this way. Further adaptation is possible (for example just to keep the mean_sd columns (just use transmute instead of mutate):
Update:
library(dplyr)
library(stringr)
df %>%
mutate(across(-c(season, Site), ~round(.,2))) %>%
mutate(across(ends_with('_1'), ~ paste0(.,
"(",
get(str_replace(cur_column(), "_1$", "_2")),
")"
), .names = "mean_sd_{.col}")) %>%
rename_at(vars(starts_with('mean_sd')), ~ str_remove(., "\\_1"))
season Site Temp_1 Temp_2 pH_1 pH_2 DO_1 DO_2 EC_1 EC_2 SS_1 SS_2 TP_1 TP_2 SRP_1 SRP_2 PP_1 PP_2 DA_1 DA_2 DNI_1 DNI_2 DNA_1 DNA_2 DF_1
1 Winter 1 7.20 1.56 7.43 0.29 9.00 0.00 337.5 55.86 5.94 0.73 73.5 3.54 19.0 2.83 54.5 6.36 0.08 0.00 0.05 0.03 20.44 1.80 0.10
2 Winter 2 7.05 1.91 7.38 0.28 9.10 0.42 333.0 41.01 3.65 1.06 75.0 12.73 19.0 1.41 56.0 11.31 0.08 0.01 0.04 0.02 20.44 1.27 0.09
3 Winter 3 6.30 1.70 7.52 0.10 8.25 0.07 321.5 51.62 5.03 2.93 61.5 9.19 10.0 2.83 51.5 6.36 0.08 0.01 0.06 0.02 14.99 2.29 0.09
4 Winter 4 6.25 2.33 7.53 0.12 8.85 0.49 322.0 32.53 2.54 0.47 66.5 6.36 14.0 0.00 52.5 6.36 0.04 0.00 0.03 0.02 19.95 1.03 0.10
5 Winter 5 6.20 2.40 7.38 0.06 9.25 0.64 309.0 25.46 10.22 8.23 83.0 26.87 13.5 0.71 69.5 26.16 0.06 0.01 0.04 0.03 17.03 0.40 0.10
6 Winter 6 5.40 2.40 7.57 0.26 9.00 0.42 300.5 30.41 4.60 0.67 87.0 24.04 23.5 3.54 63.5 20.51 0.05 0.01 0.01 0.02 8.58 1.72 0.09
DF_2 DC_1 DC_2 DS_1 DS_2 DOC_1 DOC_2 mean_sd_Temp mean_sd_pH mean_sd_DO mean_sd_EC mean_sd_SS mean_sd_TP mean_sd_SRP mean_sd_PP mean_sd_DA
1 0.00 12.19 0.58 15.83 1.67 19.74 2.74 7.2(1.56) 7.43(0.29) 9(0) 337.5(55.86) 5.94(0.73) 73.5(3.54) 19(2.83) 54.5(6.36) 0.08(0)
2 0.01 12.74 0.04 15.70 1.70 20.08 1.70 7.05(1.91) 7.38(0.28) 9.1(0.42) 333(41.01) 3.65(1.06) 75(12.73) 19(1.41) 56(11.31) 0.08(0.01)
3 0.01 13.10 0.70 14.38 2.48 21.24 2.60 6.3(1.7) 7.52(0.1) 8.25(0.07) 321.5(51.62) 5.03(2.93) 61.5(9.19) 10(2.83) 51.5(6.36) 0.08(0.01)
4 0.01 13.42 0.13 15.44 1.04 20.34 1.05 6.25(2.33) 7.53(0.12) 8.85(0.49) 322(32.53) 2.54(0.47) 66.5(6.36) 14(0) 52.5(6.36) 0.04(0)
5 0.00 15.14 0.67 14.65 1.24 21.88 0.23 6.2(2.4) 7.38(0.06) 9.25(0.64) 309(25.46) 10.22(8.23) 83(26.87) 13.5(0.71) 69.5(26.16) 0.06(0.01)
6 0.01 21.43 0.17 11.99 0.98 24.92 0.45 5.4(2.4) 7.57(0.26) 9(0.42) 300.5(30.41) 4.6(0.67) 87(24.04) 23.5(3.54) 63.5(20.51) 0.05(0.01)
mean_sd_DNI mean_sd_DNA mean_sd_DF mean_sd_DC mean_sd_DS mean_sd_DOC
1 0.05(0.03) 20.44(1.8) 0.1(0) 12.19(0.58) 15.83(1.67) 19.74(2.74)
2 0.04(0.02) 20.44(1.27) 0.09(0.01) 12.74(0.04) 15.7(1.7) 20.08(1.7)
3 0.06(0.02) 14.99(2.29) 0.09(0.01) 13.1(0.7) 14.38(2.48) 21.24(2.6)
4 0.03(0.02) 19.95(1.03) 0.1(0.01) 13.42(0.13) 15.44(1.04) 20.34(1.05)
5 0.04(0.03) 17.03(0.4) 0.1(0) 15.14(0.67) 14.65(1.24) 21.88(0.23)
6 0.01(0.02) 8.58(1.72) 0.09(0.01) 21.43(0.17) 11.99(0.98) 24.92(0.45)
First answer: We could do this like so:
library(dplyr)
df %>% mutate(mean_sd = paste0(Temp_1, " (", round(Temp_2,2), ")"), .before=5)
season Site Temp_1 Temp_2 mean_sd pH_1 pH_2 DO_1 DO_2 EC_1 EC_2 SS_1 SS_2 TP_1 TP_2 SRP_1 SRP_2 PP_1
1 Winter 1 7.20 1.555635 7.2 (1.56) 7.435 0.28991378 9.00 0.00000000 337.5 55.86144 5.945 0.7283200 73.5 3.535534 19.0 2.8284271 54.5
2 Winter 2 7.05 1.909188 7.05 (1.91) 7.380 0.28284271 9.10 0.42426407 333.0 41.01219 3.650 1.0606602 75.0 12.727922 19.0 1.4142136 56.0
3 Winter 3 6.30 1.697056 6.3 (1.7) 7.520 0.09899495 8.25 0.07071068 321.5 51.61880 5.025 2.9344931 61.5 9.192388 10.0 2.8284271 51.5
4 Winter 4 6.25 2.333452 6.25 (2.33) 7.525 0.12020815 8.85 0.49497475 322.0 32.52691 2.535 0.4737615 66.5 6.363961 14.0 0.0000000 52.5
5 Winter 5 6.20 2.404163 6.2 (2.4) 7.380 0.05656854 9.25 0.63639610 309.0 25.45584 10.220 8.2307229 83.0 26.870058 13.5 0.7071068 69.5
6 Winter 6 5.40 2.404163 5.4 (2.4) 7.565 0.26162951 9.00 0.42426407 300.5 30.40559 4.595 0.6717514 87.0 24.041631 23.5 3.5355339 63.5
PP_2 DA_1 DA_2 DNI_1 DNI_2 DNA_1 DNA_2 DF_1 DF_2 DC_1 DC_2 DS_1 DS_2 DOC_1
1 6.363961 0.0830 0.002828427 0.04873944 0.03450791 20.43508 1.8028811 0.09923797 0.003720723 12.18685 0.5761588 15.83438 1.6715314 19.74
2 11.313708 0.0775 0.012020815 0.04101556 0.02233125 20.43892 1.2687128 0.09472684 0.007249143 12.73924 0.0430072 15.69623 1.6997877 20.08
3 6.363961 0.0775 0.009192388 0.06177234 0.01873602 14.98693 2.2883937 0.09432312 0.014293247 13.09550 0.7025396 14.37636 2.4756057 21.24
4 6.363961 0.0440 0.001414214 0.03374418 0.01620325 19.95341 1.0311617 0.09806487 0.011689547 13.41756 0.1346425 15.44424 1.0383126 20.34
5 26.162951 0.0590 0.012727922 0.04148094 0.02581691 17.03060 0.3965285 0.09803047 0.002556718 15.14098 0.6678661 14.64760 1.2448876 21.88
6 20.506097 0.0510 0.008485281 0.01434617 0.02028854 8.57675 1.7235083 0.08578391 0.008305191 21.42976 0.1701289 11.98774 0.9754832 24.92
DOC_2
1 2.7435743
2 1.6970563
3 2.6021530
4 1.0465180
5 0.2262742
6 0.4525483
CodePudding user response:
You can create a new column like this
df$Temp <- paste0(df$Temp_1, ' (', df$Temp_2, ')')
And select only the desired output columns
df[, c('season', 'Site', 'Temp')]
CodePudding user response:
library(tidyverse)
df %>%
pivot_longer(-c(season, Site)) %>%
mutate(name = name %>% str_remove_all("[^a-zA-Z]")) %>%
group_by(season, Site, name) %>%
summarise(value = str_c(round(value, 2), collapse = ", ")) %>%
pivot_wider(names_from = name,
values_from = value)
# A tibble: 6 x 17
# Groups: season, Site [6]
season Site DA DC DF DNA DNI DO DOC DS EC pH PP SRP SS Temp TP
<fct> <fct> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 Winter 1 0.08, 0 12.19, 0.58 0.1, 0 20.44, 1.8 0.05, 0.03 9, 0 19.7~ 15.8~ 337.~ 7.43~ 54.5~ 19, ~ 5.94~ 7.2,~ 73.5~
2 Winter 2 0.08, 0.01 12.74, 0.04 0.09, 0.01 20.44, 1.27 0.04, 0.02 9.1, 0.~ 20.0~ 15.7~ 333,~ 7.38~ 56, ~ 19, ~ 3.65~ 7.05~ 75, ~
3 Winter 3 0.08, 0.01 13.1, 0.7 0.09, 0.01 14.99, 2.29 0.06, 0.02 8.25, 0~ 21.2~ 14.3~ 321.~ 7.52~ 51.5~ 10, ~ 5.03~ 6.3,~ 61.5~
4 Winter 4 0.04, 0 13.42, 0.13 0.1, 0.01 19.95, 1.03 0.03, 0.02 8.85, 0~ 20.3~ 15.4~ 322,~ 7.53~ 52.5~ 14, 0 2.54~ 6.25~ 66.5~
5 Winter 5 0.06, 0.01 15.14, 0.67 0.1, 0 17.03, 0.4 0.04, 0.03 9.25, 0~ 21.8~ 14.6~ 309,~ 7.38~ 69.5~ 13.5~ 10.2~ 6.2,~ 83, ~
6 Winter 6 0.05, 0.01 21.43, 0.17 0.09, 0.01 8.58, 1.72 0.01, 0.02 9, 0.42 24.9~ 11.9~ 300.~ 7.57~ 63.5~ 23.5~ 4.6,~ 5.4,~ 87, ~