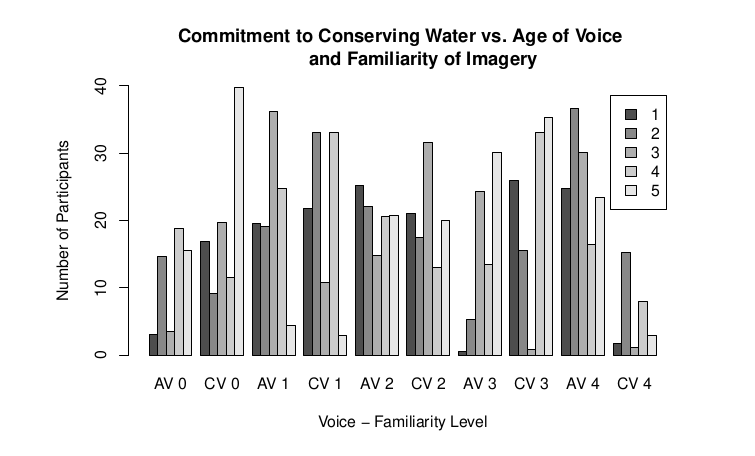
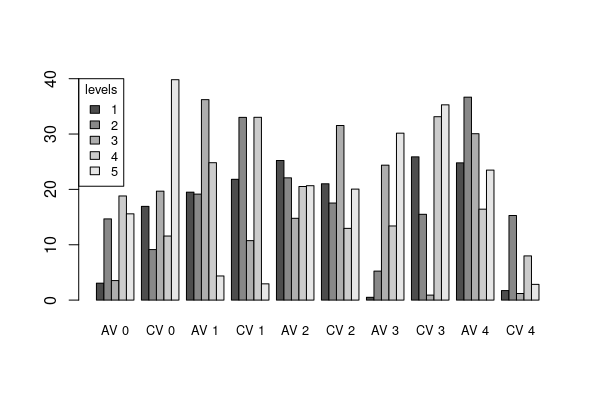
So I'm trying to make a barplot on R that has a categorical label, not a number scale, as the x-axis. I then need to knit the RMarkdown file as a pdf. This is what it looks like before I knit it as a RMD:
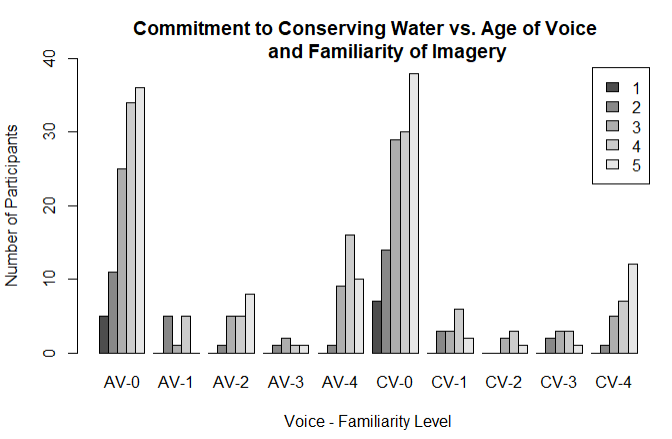
This is what it looks like after I knit it as an RMD:
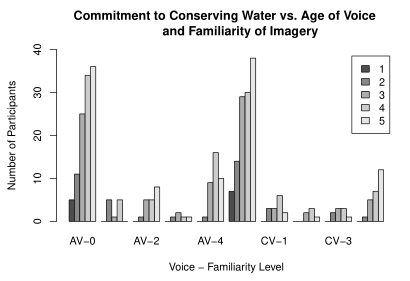
This is the code for the plot (I know there are no issues with this code for my purposes; I'm just including it so that stack overflow doesn't think I'm spamming):
`
barplot(t1,
legend = TRUE,
main="Commitment to Conserving Water vs. Age of Voice
and Familiarity of Imagery",
xlab = "Voice - Familiarity Level",
ylab = "Number of Participants",
ylim = c(0,40),
beside = TRUE)
`
As you can see in the images, hopefully, I lose some of the labels on the x-axis when I knit into RMD. I think this is because the x labels are too long, so R is just cutting every other one out to make the graph look neater. I either need a way to prevent R from doing this, or to make the x labels smaller.
This is a school assignment where I can't use ggplot2 or anything like that; I have to do this in base R. I tried using cex.axis, but for some reason it would only change the size of the y axis font and not the x axis font, which might be because there's not really an axis for x. I also tried looking into rotating the x axis labels, if I can't make it smaller, but I couldn't find out how to do that in base R.
CodePudding user response:
As already stated in comments, just play around with fig.width=, fig.height= chunk parameters. I chose a 6:4 ratio because it's standard in some journals.
---
output: pdf_document
---
```{r, echo=FALSE, fig.width=7, fig.height=4.67}
## simulate some data
set.seed(580509)
t1 <- replicate(10, runif(5, 0, 40)) |> `dimnames<-`(list(1:5, outer(c('AV', 'CV'), 0:4, paste)))
## plot
barplot(t1,
legend=TRUE,
main="Commitment to Conserving Water vs. Age of Voice
and Familiarity of Imagery",
xlab="Voice - Familiarity Level",
ylab="Number of Participants",
ylim=c(0, 40),
beside=TRUE)
```
To change font size of "x-axis" you want to use the cex.names= argument. This is also possible for the legend; using the args.legend= argument you may specify all arguments of the ?legend function.
## sim. data
set.seed(580509)
t1 <- replicate(10, runif(5, 0, 40)) |> `dimnames<-`(list(1:5, outer(c('AV', 'CV'), 0:4, paste)))
barplot(t1, legend=TRUE, ylim=c(0, 40), beside=TRUE,
cex.names=.8,
args.legend=list(x='topleft', cex=.8, title='levels')
)
Study the help ?barplot carefully; it is really a powerful function, great your lecturer teaches it! It is IMO superior to ggplot, because it is able to directly process matrices, looks more professional, and is more intuitive.
CodePudding user response:
Welcome to stackoverflow. You are knitting it into R Markdown document. Have a look at the code chunk, which contains this code. I think currently it looks like
{r}
barplot(t1,
legend = TRUE,
main="Commitment to Conserving Water vs. Age of Voice
and Familiarity of Imagery",
xlab = "Voice - Familiarity Level",
ylab = "Number of Participants",
ylim = c(0,40),
beside = TRUE)
Change the first line only.
{r fig.width=7}
barplot(t1,
legend = TRUE,
main="Commitment to Conserving Water vs. Age of Voice
and Familiarity of Imagery",
xlab = "Voice - Familiarity Level",
ylab = "Number of Participants",
ylim = c(0,40),
beside = TRUE)
You can experiment with different fig.width number to see how it looks like in the final document.