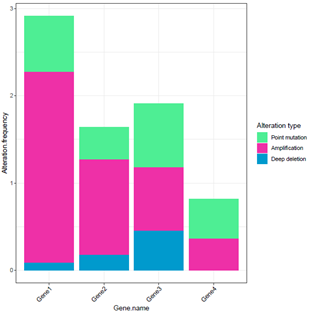
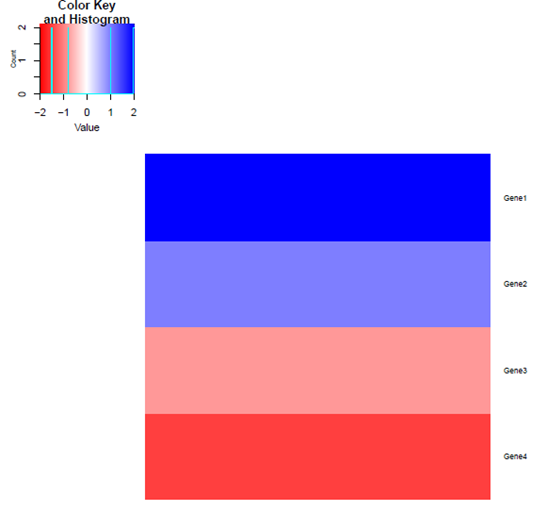
I have these two plots (a heatmap and a stacked barplot). How can I put these plots near each other? Indeed I want to integrate these two plots.
Thanks for any help.
Here is my script:
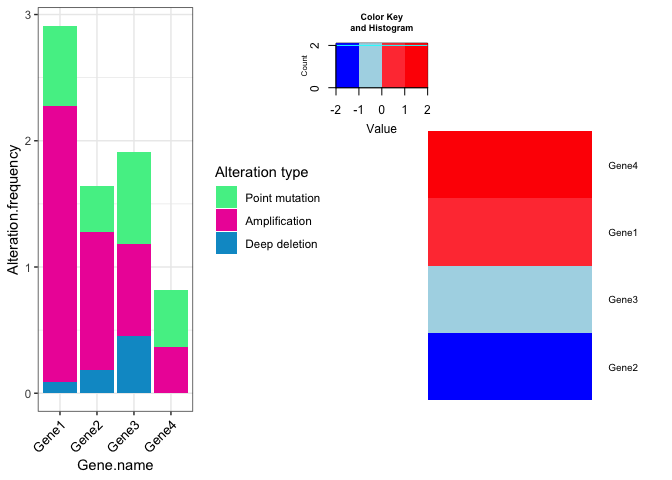
df.bar <- data.frame(Gene.name=c('Gene1','Gene1','Gene1','Gene2','Gene2','Gene2','Gene3','Gene3','Gene3',
'Gene4','Gene4','Gene4'),
Alteration.frequency=c(0.0909, 2.1838, 0.6369, 0.1819, 1.0919, 0.3639, 0.4549,0.7279,
0.7279, 0.000, 0.3639, 0.4549),
Alteration.Type=c("Deep deletion", "Amplification", "Point mutation", "Deep deletion",
"Amplification", "Point mutation", "Deep deletion", "Amplification" ,
"Point mutation", "Deep deletion", "Amplification" , "Point mutation"))
library(ggplot2)
ggplot(df.bar,
aes(fill=factor(Alteration.Type, levels = c('Point mutation','Amplification','Deep deletion')),
y=Alteration.frequency, x=Gene.name))
geom_bar(position="stack", stat="identity") theme_bw()
theme(axis.text.x = element_text(size = 10, angle = 45, hjust = 1, colour = 'black'))
scale_fill_manual(values=c("seagreen2", "maroon2",'deepskyblue3'))
labs(fill = 'Alteration type')
df.heatmap <- data.frame(Gene_name=c('Gene1','Gene2','Gene3','Gene4'),
log2FC=c(0.56,-1.5,-0.8,2))
library(gplots)
heatmap.2(cbind(df.heatmap$log2FC, df.heatmap$log2FC), trace="n", Colv = NA,
dendrogram = "none", labCol = "", labRow = df$Gene_name, cexRow = 0.75,
col=my_palette)
I need such this plot.
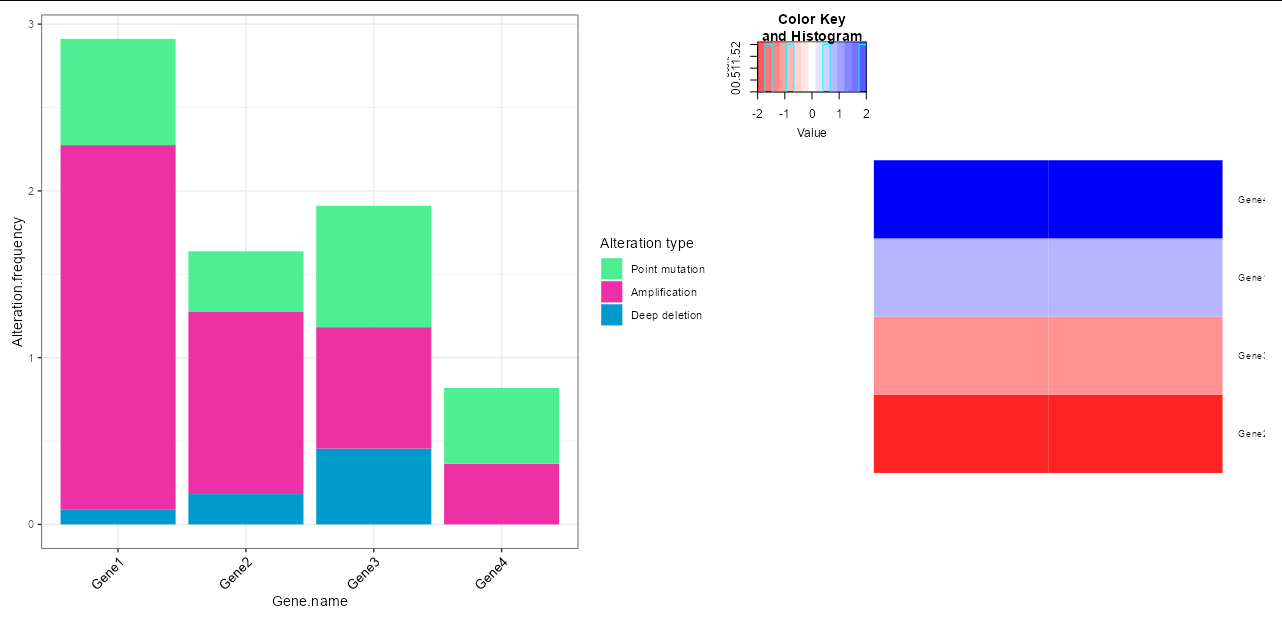
CodePudding user response:
There are various ways to mix base R graphics like heatmap.2 and grid-based graphics like ggplot, but this case is a little more complex due to the fact that heatmap.2 seems to use multiple plotting regions. However, the following solution should do the trick:
library(gplots)
library(ggplot2)
library(patchwork)
library(gridGraphics)
p1 <- ggplot(df.bar,
aes(fill = factor(Alteration.Type, levels = c('Point mutation',
'Amplification',
'Deep deletion')),
y = Alteration.frequency, x = Gene.name))
geom_col(position = "stack")
theme_bw()
theme(axis.text.x = element_text(size = 10, angle = 45, hjust = 1,
colour = 'black'))
scale_fill_manual(values = c("seagreen2", "maroon2", 'deepskyblue3'))
labs(fill = 'Alteration type')
heatmap.2(cbind(df.heatmap$log2FC, df.heatmap$log2FC), trace = "n",
Colv = NA, dendrogram = "none", labCol = "",
labRow = df.heatmap$Gene_name, cexRow = 0.75,
col = colorRampPalette(c("red", "white", "blue")))
grid.echo()
p2 <- wrap_elements(grid.grab())
p1 p2
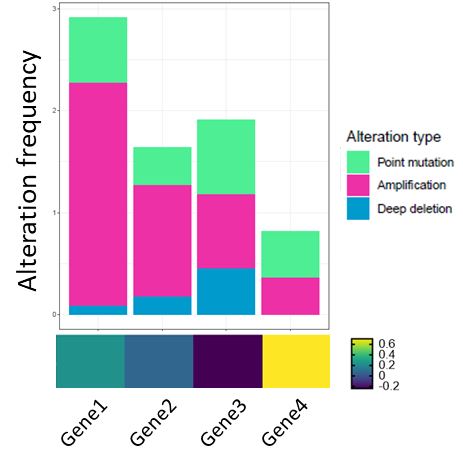
CodePudding user response:
Another option could be using plot_grid from the package cowplot to combine ggplot and basegraphics or other framework. For the heatmap you could use ~ and I slightly modified the size of the legend to fit it better. Here is a reproducible example:
library(ggplot2)
library(cowplot)
library(gridGraphics)
library(gplots)
# first plot
p1 <- ggplot(df.bar,
aes(fill=factor(Alteration.Type, levels = c('Point mutation','Amplification','Deep deletion')),
y=Alteration.frequency, x=Gene.name))
geom_bar(position="stack", stat="identity") theme_bw()
theme(axis.text.x = element_text(size = 10, angle = 45, hjust = 1, colour = 'black'))
scale_fill_manual(values=c("seagreen2", "maroon2",'deepskyblue3'))
labs(fill = 'Alteration type')
# Second plot
p2<- ~heatmap.2(cbind(df.heatmap$log2FC, df.heatmap$log2FC), trace="n", Colv = NA,
dendrogram = "none", labCol = "", labRow = df.heatmap$Gene_name, cexRow = 0.75,
col=my_palette, key.par=list(mar=c(3,0,3,0), cex.main = 0.75), keysize = 1.5)
# combine all plots together
plot_grid(p1, p2)
Created on 2022-12-03 with reprex v2.0.2