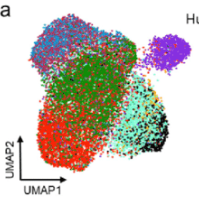
More often I see in publications that instead of printing the UMAP axis in scRNAseq experiments (or even t-SNE or PCA) they just add two small arrows in the bottom left corner. Something like this:
I really like the aesthetics of it but I don´t know how to replicate this in R. I guess this is normally done separately with some image editor but it can probably be done with ggplot2 package to make it more reproducible.
So far I only got the arrows in the axis:
x <- data.frame(UMAP1=rnorm(300),UMAP2=rnorm(300))
ggplot(x, aes(UMAP1,UMAP2)) geom_point() theme_minimal()
theme(axis.line = element_line(arrow = arrow(type='closed',
length = unit(10,'pt'))))
But I don't know how to make them smaller and with the title underneath. Does anyone have any suggestions on how to do this?
CodePudding user response:
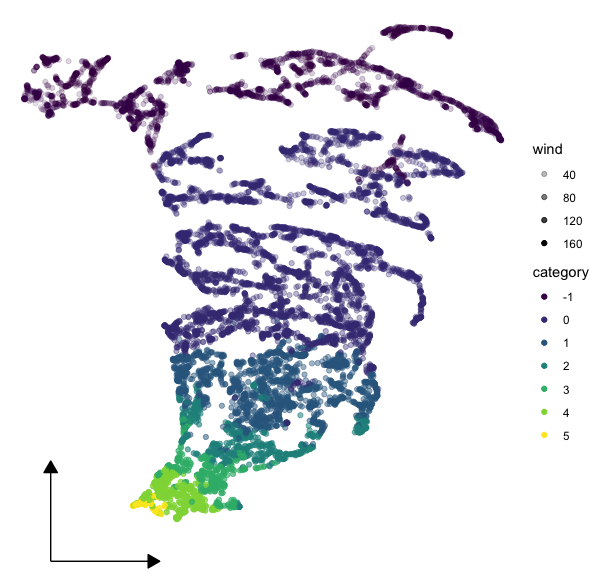
I'd suggest faking it with an annotation:
library(dplyr); library(umap); library(ggplot2)
arr <- list(x = -10, y = -15, x_len = 5, y_len = 5)
ggplot(storms_umap_extract, aes(x,y, color = category, alpha = wind))
geom_point()
annotate("segment",
x = arr$x, xend = arr$x c(arr$x_len, 0),
y = arr$y, yend = arr$y c(0, arr$y_len),
arrow = arrow(type = "closed", length = unit(10, 'pt')))
theme_void()
Here's the umap data:
storms_umap <- storms |>
select(lat,long, wind, pressure) |>
umap() # this took about a minute to run
storms_umap_extract <- tibble(
x = storms_umap$layout[,1],
y = storms_umap$layout[,2],
wind = storms_umap$data[,3],
category = storms$category
)
CodePudding user response:
In code below, adjust unit(3, "cm") and hjust = 0 to taste.
Disclaimer: I wrote ggh4x.
library(ggplot2)
axis <- ggh4x::guide_axis_truncated(
trunc_lower = unit(0, "npc"),
trunc_upper = unit(3, "cm")
)
x <- data.frame(UMAP1=rnorm(300),UMAP2=rnorm(300))
ggplot(x, aes(UMAP1, UMAP2))
geom_point()
guides(x = axis, y = axis)
theme(axis.line = element_line(arrow = arrow()),
axis.title = element_text(hjust = 0))
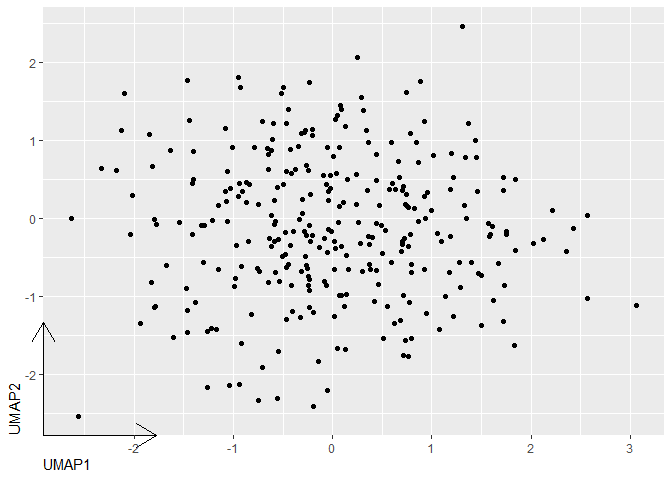
Created on 2022-12-07 by the reprex package (v2.0.1)
Optionally, add the code below if you want to get rid of the ticks and labels (which don't make any sense in terms of UMAP/tSNE anyway)
scale_x_continuous(breaks = NULL)
scale_y_continuous(breaks = NULL)