Here is an reproducible data example
library(data.table)
testDT = fread(
"from to
Root A
A x1
x1 y1
y1 z1
y1 B
Root B
B x3
x3 y2
Root C")
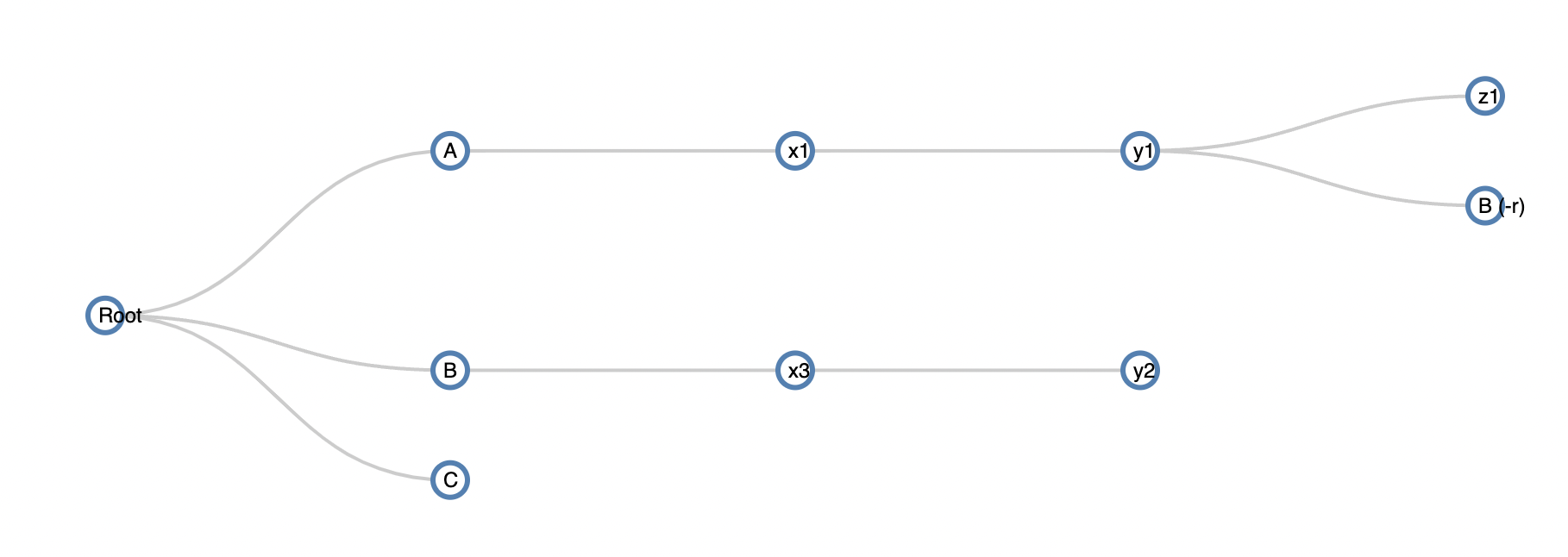
An example visualization of the data:
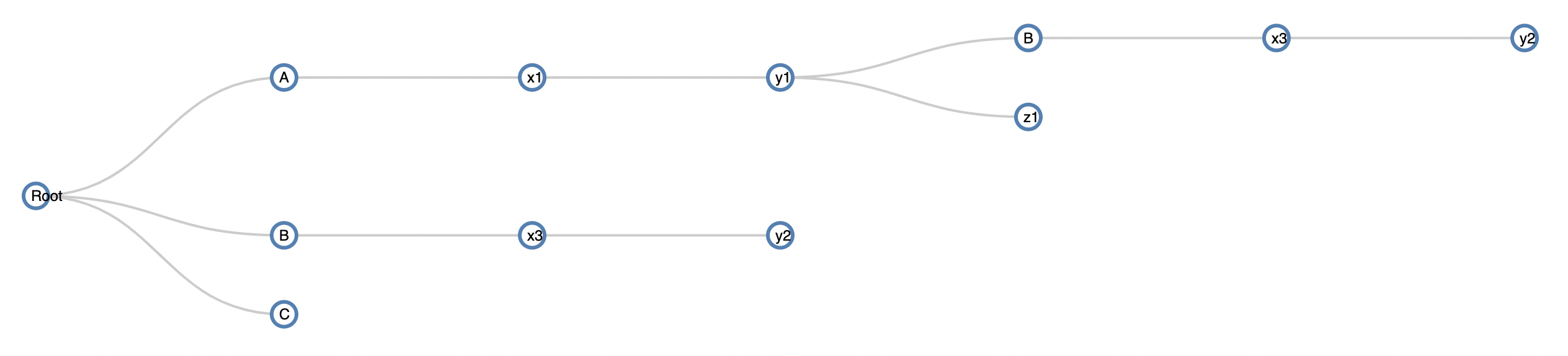
As you see, hierarchically, the same node B at level1 appears as a child of node y1. What I would like to do is remove if the repeating node B has children and tag it. The output I want is:
The output table that i desired:
| from | to |
|-------------|---------------|
| Root | A |
| A | x1 |
| x1 | y1 |
| y1 | z1 |
| y1 | B (-r) |
| Root | B |
| B | x3 |
| x3 | y2 |
| Root | C |
I had prepared a question to remove the node that caused the recursive build and they provided a solution thanks. However, this solution completely deletes node B connected to y1 against my will.
CodePudding user response:
I couldn't come up with a simple igraph solution as all the traverse functions in igraph will not visit the same node twice. Therefore I wrote a bfs algorithm that will keep track of edges where the child has already been visited.
library(purrr)
library(igraph)
library(dplyr)
G <- graph_from_data_frame(testDT)
stack <- 'Root'
traversed <- c()
edges <- data.frame(from = c(), to = c(), traversed = c())
while(length(stack) > 0) {
el <- stack[1]
stack <- stack[-1]
children <- adjacent_vertices(G, el, mode = "out" )[[1]] |>
map_chr(~names(V(G)[.x]))
if(!el %in% traversed) {
traversed <- c(el, traversed)
if (length(children) > 0) {
edges_ <- data.frame(from = el, to = children) |>
mutate(traversed = to %in% traversed))
stack <- c(stack, edges_ |> filter(!traversed) |> pull(to))
edges <- rbind(edges_, edges)
}
}
}
edges
##> from to traversed
##> B y1 B TRUE
##> z1 y1 z1 FALSE
##> y2 x3 y2 FALSE
##> y1 x1 y1 FALSE
##> x3 B x3 FALSE
##> x1 A x1 FALSE
##> A Root A FALSE
##> B1 Root B FALSE
##> C Root C FALSE
Then, with simple data manipulation you can obtain the desired format.
edges |>
mutate(to = ifelse(traversed,
paste(to, "(-r)"),
to),
.keep = "unused")
##> from to
##> B y1 B (-r)
##> z1 y1 z1
##> y2 x3 y2
##> y1 x1 y1
##> x3 B x3
##> x1 A x1
##> A Root A
##> B1 Root B
##> C Root C