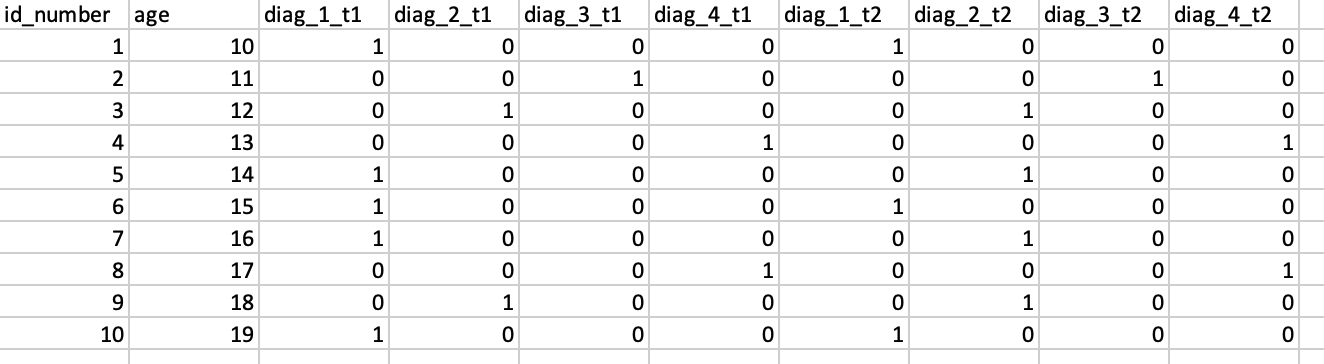
I have a dataset df where diag_t1 = diagnosis for time1, diag_t2 = diagnosis for time2, diag_t3 = diagnosis for time3 and diag_t4 = diagnosis for time4. There are four possible diagnosis = 1,2,3 and 4. 0 is when there is no diagnosis.
df <- data.frame(
id_number = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10),
age = c(10, 11, 12, 13, 14, 15, 16, 17, 18, 19),
diag_t1 = c(1, 3, 2, 4, 1,1 , 1, 4, 2, 1),
diag_t2 = c(1, 3, 2, 4, 2, 1, 2, 4, 2, 1),
diag_t3 = c(1, 3, 2, 3, 2, 1, 2, 4, 2, 1),
diag_t4 = c(0, 3, 2, 3, 2, 1, 2, 4, 2, 1)
)
I am trying to change this in a format where I have all one column for each disorder for each time. This is the output I want. Can anyone help on this? Thank-you
CodePudding user response:
With fastDummies::dummy_cols:
library(dplyr)
library(fastDummies)
df %>%
mutate(across(starts_with("diag"), factor, levels = c(0:4))) %>%
dummy_cols(remove_selected_columns = TRUE, remove_first_dummy = TRUE)
output
id_number age diag_t1_1 diag_t1_2 diag_t1_3 diag_t1_4 diag_t2_1 diag_t2_2 diag_t2_3 diag_t2_4 diag_t3_1 diag_t3_2 diag_t3_3
1 1 10 1 0 0 0 1 0 0 0 1 0 0
2 2 11 0 0 1 0 0 0 1 0 0 0 1
3 3 12 0 1 0 0 0 1 0 0 0 1 0
4 4 13 0 0 0 1 0 0 0 1 0 0 1
5 5 14 1 0 0 0 0 1 0 0 0 1 0
6 6 15 1 0 0 0 1 0 0 0 1 0 0
7 7 16 1 0 0 0 0 1 0 0 0 1 0
8 8 17 0 0 0 1 0 0 0 1 0 0 0
9 9 18 0 1 0 0 0 1 0 0 0 1 0
10 10 19 1 0 0 0 1 0 0 0 1 0 0
diag_t3_4 diag_t4_1 diag_t4_2 diag_t4_3 diag_t4_4
1 0 0 0 0 0
2 0 0 0 1 0
3 0 0 1 0 0
4 0 0 0 1 0
5 0 0 1 0 0
6 0 1 0 0 0
7 0 0 1 0 0
8 1 0 0 0 1
9 0 0 1 0 0
10 0 1 0 0 0