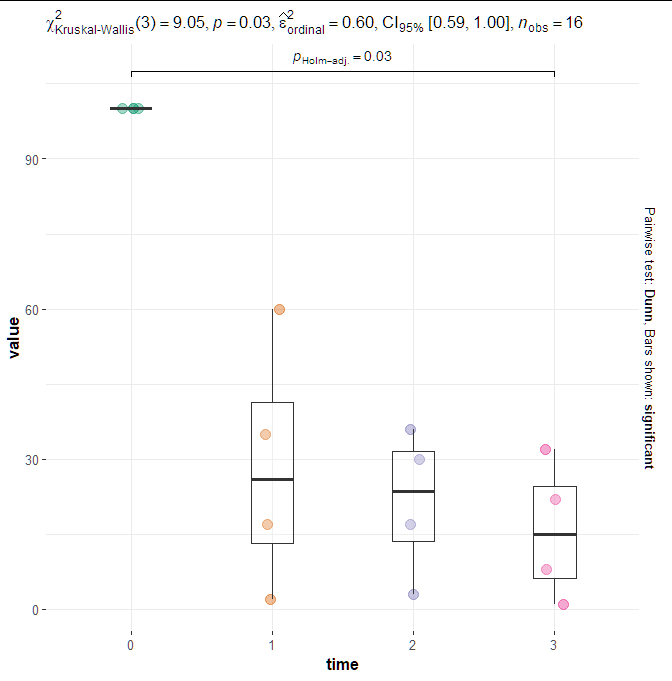
I am trying to use grouped_ggwithinstats() function from ggstatsplot package:
I have this data set:
df <- structure(list(time = c(0L, 1L, 2L, 3L, 0L, 1L, 2L, 3L, 0L, 1L,
2L, 3L, 0L, 1L, 2L, 3L), group1 = c("A", "A", "A", "A", "B",
"B", "B", "B", "C 1", "C 1", "C 1", "C 1", "C 2", "C 2", "C 2",
"C 2"), group2 = c("Z", "Z", "Z", "Z", "Z", "Z", "Z", "Z", "Z",
"Z", "Z", "Z", "Z", "Z", "Z", "Z"), value = c(100L, 60L, 30L,
32L, 100L, 2L, 3L, 1L, 100L, 17L, 17L, 8L, 100L, 35L, 36L, 22L
)), class = "data.frame", row.names = c("1", "2", "3", "4", "5",
"6", "7", "8", "9", "10", "11", "12", "13", "14", "15", "16"))
time group1 group2 value
1 0 A Z 100
2 1 A Z 60
3 2 A Z 30
4 3 A Z 32
5 0 B Z 100
6 1 B Z 2
7 2 B Z 3
8 3 B Z 1
9 0 C 1 Z 100
10 1 C 1 Z 17
11 2 C 1 Z 17
12 3 C 1 Z 8
13 0 C 2 Z 100
14 1 C 2 Z 35
15 2 C 2 Z 36
16 3 C 2 Z 22
Using this code:
library(ggstatsplot)
ggbetweenstats(
data = df,
x = time,
y = value,
type = "nonparametric",
plot.type = "box",
centrality.plotting = FALSE
)
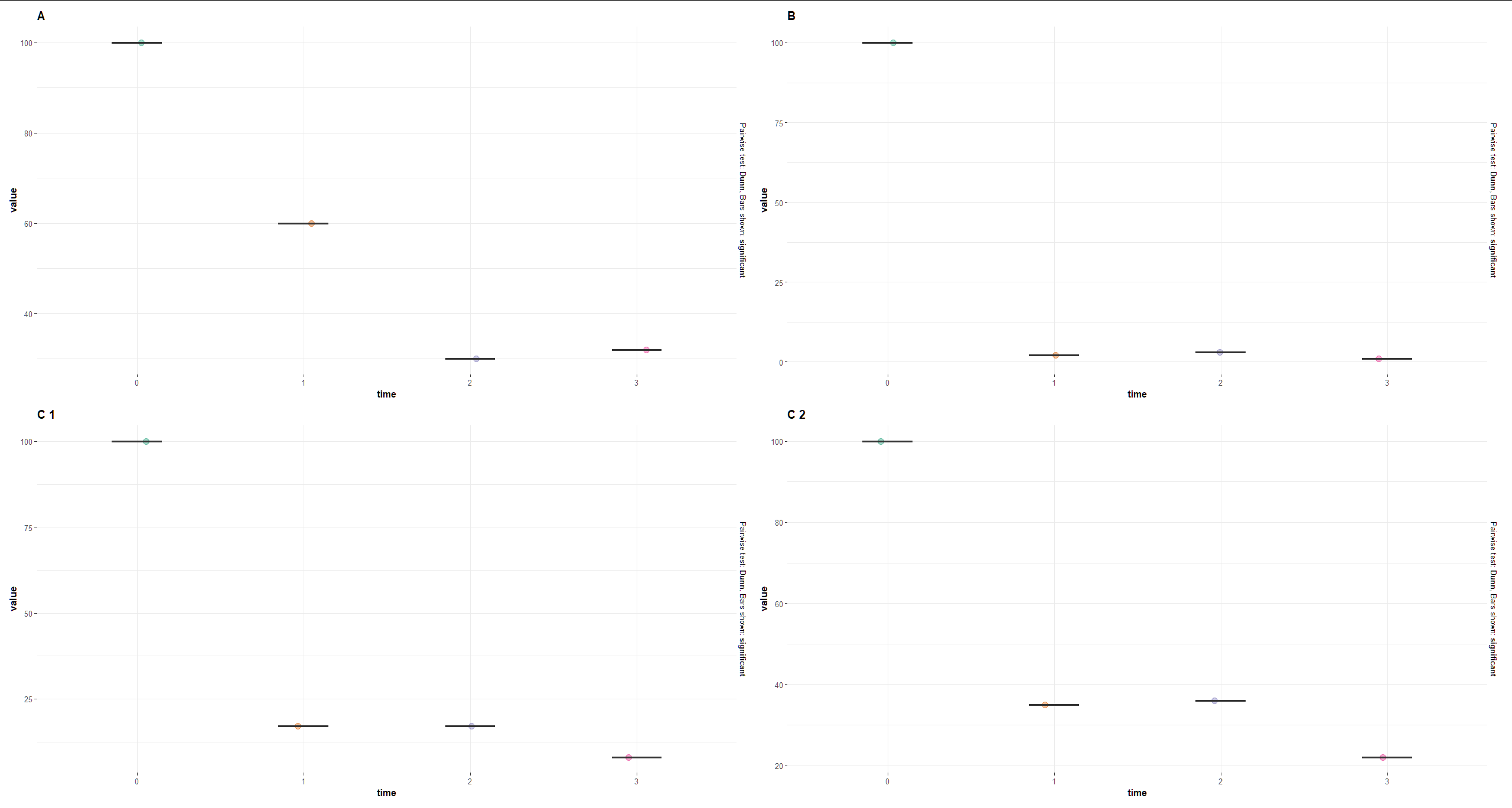
 I now would like to group by
I now would like to group by group1 and mark the dots with each group:
So when I do:
grouped_ggbetweenstats(
data = df,
x = time,
y = value,
grouping.var = group1,
type = "nonparametric",
plot.type = "box",
centrality.plotting = FALSE
)
I do not understand the difference between ggwithinstats vs. ggbetweenstats in relation to grouped_ggbetweenstats() and grouped_ggwithinstats() the latter giving the Error:
Error in data.frame(..., check.names = FALSE) :
arguments imply differing number of rows: 0, 1
What I try to test is using repeated anova, if the changes over timepoint 0,1,2,3 are significant within each group. e.g. in GroupA, Group B, Group C 1, Group C 2.
CodePudding user response:
You are using grouped_ggwithinstats() correctly, but you are getting all these warnings because there just isn't enough data to run hypothesis testing and estimation.
You have one datum per condition per group, and so no statistical tests can be run, and thus the subtitles and captions are empty.
library(dplyr, warn.conflicts = FALSE)
library(ggstatsplot)
#> You can cite this package as:
#> Patil, I. (2021). Visualizations with statistical details: The 'ggstatsplot' approach.
#> Journal of Open Source Software, 6(61), 3167, doi:10.21105/joss.03167
df <- tibble(time = c(
0L, 1L, 2L, 3L, 0L, 1L, 2L, 3L, 0L, 1L,
2L, 3L, 0L, 1L, 2L, 3L
), group1 = c(
"A", "A", "A", "A", "B",
"B", "B", "B", "C 1", "C 1", "C 1", "C 1", "C 2", "C 2", "C 2",
"C 2"
), group2 = c(
"Z", "Z", "Z", "Z", "Z", "Z", "Z", "Z", "Z",
"Z", "Z", "Z", "Z", "Z", "Z", "Z"
), value = c(
100L, 60L, 30L,
32L, 100L, 2L, 3L, 1L, 100L, 17L, 17L, 8L, 100L, 35L, 36L, 22L
))
df %>%
group_by(group1, time) %>%
count()
#> # A tibble: 16 × 3
#> # Groups: group1, time [16]
#> group1 time n
#> <chr> <int> <int>
#> 1 A 0 1
#> 2 A 1 1
#> 3 A 2 1
#> 4 A 3 1
#> 5 B 0 1
#> 6 B 1 1
#> 7 B 2 1
#> 8 B 3 1
#> 9 C 1 0 1
#> 10 C 1 1 1
#> 11 C 1 2 1
#> 12 C 1 3 1
#> 13 C 2 0 1
#> 14 C 2 1 1
#> 15 C 2 2 1
#> 16 C 2 3 1
grouped_ggwithinstats(
data = df,
x = time,
y = value,
grouping.var = group1,
type = "nonparametric",
pairwise.comparisons = FALSE,
centrality.plotting = FALSE
)
#> Warning: Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Warning in max(data$density): no non-missing arguments to max; returning -Inf
#> Warning: Computation failed in `stat_ydensity()`
#> Caused by error in `$<-.data.frame`:
#> ! replacement has 1 row, data has 0
#> Warning: Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Warning in max(data$density): no non-missing arguments to max; returning -Inf
#> Warning: Computation failed in `stat_ydensity()`
#> Caused by error in `$<-.data.frame`:
#> ! replacement has 1 row, data has 0
#> Warning: Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Warning in max(data$density): no non-missing arguments to max; returning -Inf
#> Warning: Computation failed in `stat_ydensity()`
#> Caused by error in `$<-.data.frame`:
#> ! replacement has 1 row, data has 0
#> Warning: Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Groups with fewer than two data points have been dropped.
#> Warning in max(data$density): no non-missing arguments to max; returning -Inf
#> Warning: Computation failed in `stat_ydensity()`
#> Caused by error in `$<-.data.frame`:
#> ! replacement has 1 row, data has 0
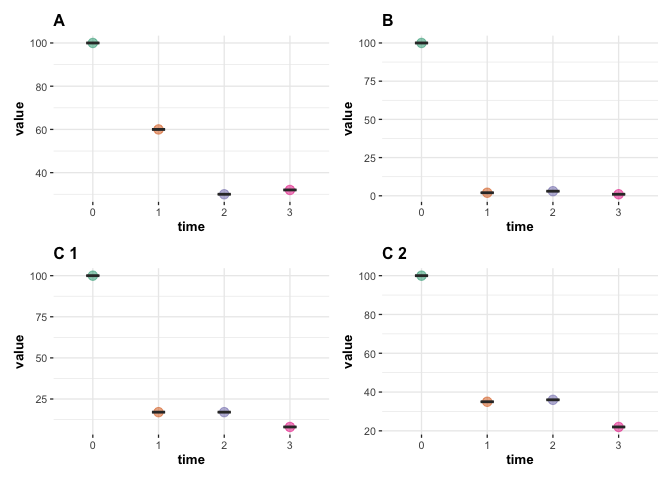
Created on 2022-12-12 with reprex v2.0.2