While I am building a networkx graph, I algorithmically sometimes add a custcol attribute only to a few of them such as:
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
G.add_edges_from([
('A','B'),
('B','C'),
('C','D'),
('D','E'),
('F','B'),
('B','G'),
('B','D'),
])
# in real life the following would be an algorithm deciding if the node
# should be custom coloured, and which colour it should get
G.nodes['C']['custcol'] = 'red' # simple setting for the example
# now let's explore the created example nodes
for node in G.nodes(data=True):
print(node)
which would print out:
('A', {})
('B', {})
('C', {'custcol': 'red'})
('D', {})
('E', {})
('F', {})
('G', {})
I am now displaying the network with a single draw such as:
NXDOPTS = {
"node_color": "orange",
"edge_color": "powderblue",
"node_size": 400,
"width": 2,
}
nx.draw(G, with_labels=True, **NXDOPTS)
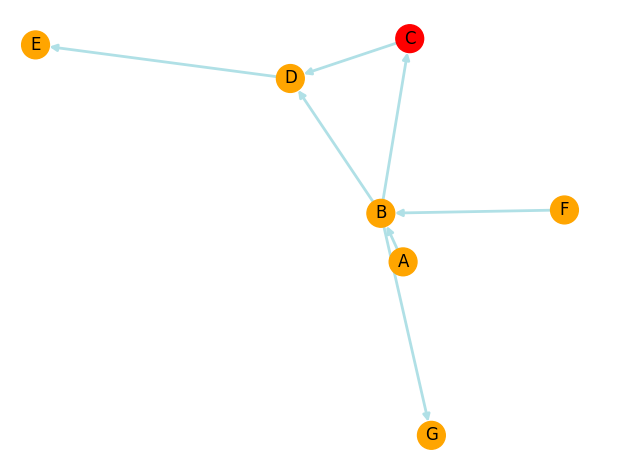
which would generate the following picture:
 What would be the best/pythonic/more efficient way of drawing node 'C' (in this example) with the colour of its custcol attribute? Of course this in reality will need to be applied to a few dozen nodes with a few different colours that are decided case by base when creating them.
What would be the best/pythonic/more efficient way of drawing node 'C' (in this example) with the colour of its custcol attribute? Of course this in reality will need to be applied to a few dozen nodes with a few different colours that are decided case by base when creating them.
CodePudding user response:
You can loop through the node data, and create a list of corresponding colors:
NXDOPTS = {
"node_color": [data["custcol"] if "custcol" in data else "orange" for _, data in G.nodes(data=True)],
"edge_color": "powderblue",
"node_size": 400,
"width": 2,
}
nx.draw(G, with_labels=True, **NXDOPTS)