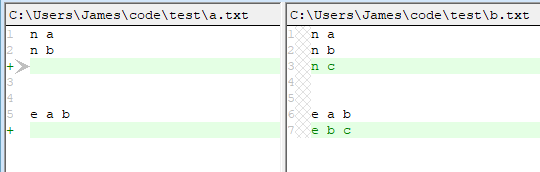
To me, this seems reasonably "meaningful".
Your use case, as you have described it, seems not to need anything more complex than this.
Here is your description
If the family tree had a version of the family in the 1990's and one in 2010's, how could I extend the graph considering v1990 and v2010?
When working with a specific version, can I isolate or exclusively use that version's data in my computations?
How is serialization affected?
In my proposal:
The graph is not extended. The versioning is done by a version control package such as GIT
You check out the required version from the version control package
You will need to design the serialization format with some care so that the differences will be most meaningful to you. You can try using a serialization function provided by the library, but will likely need to write your own to get the maximum "meaning".
If you have any more requirements than these that necessitate anything more complex, then you will need to to specify them in your question
CodePudding user response:
I would indeed reckon the data structure you are looking for is a versioning tree. E.g. one where nodes can be shared COW.
I'm not aware of things like this in the boost library.
But you can create your own data structures that have these properties. I have created one before for this answer: Transforming trees in C (look for CowTree).
Now, I'd be happy to draft something. However, your requirements are essential to the choices.
What trees would you need to be able to represent? I mean, are family trees expected to be strict DAG? That should be a safe assumption for biological family relations, but as soon as you include non-biological relations you could require more flexibility.
What kind of mutations exist to go from Treeᵥ₁ to Treeᵥ₂? If they are additive (again, as in history of biological relations) then you might be able to leverage graph adaptors:
- subgraph, where previous versions are subgraphs of later versions (the overhead would grow linear with the number of versions)
- filtered_graph, where you could use a "version timestamp" value with each node in the tree; your graph at a version (t) could then a view of the most recent version filtered for
timestamp <= t. The big benefit would be that the overhead is constant regardless of the number of versions.
I have many examples of using either adaptor in existing StackOverflow answers, in case you want to get more inspiration.
I'll mull the options while making dinner. If I think of useful demo of either approach I'll add it tonight.
Added
I just thought of the idea to represent a graph as an append-only edge list; this would be a good match if the relations need to be versioned over time, relations never (or rarely) disappear and the contents of vertices themselves need not be versioned.
As you can see, for the example code this might be an excellent match, depending perhaps on the efficiency requirements on certain algorithms. To be future-requirement safe I wouldn't jump to this appraoch, but it may have the most merit until now.