I have been working with D3 and ObservableHQ to create a Tree Plot of some hierarchical data (SARS-CoV-2 Variant lineages). I'm looking at ways to expand what I can do with the dataset and so I am moving it to Python to experiment.
One thing I did with D3 was the use d3.stratify on my tsv dataset. I am trying to recreate that data structure using python dict
class Lineage:
tree = dict()
def __init__(self, designation_date: str, pango: str, partial: str, unaliased: str):
self.designation_date = designation_date
self.pango = pango
self.partial = partial
self.unaliased = unaliased
self.parent = Lineage.parent(partial)
@classmethod
def parent(cls, lineage: str) -> Union[str, None]:
if "." in lineage:
return ".".join(lineage.split('.')[:-1])
return None
Here is the tree specific code from the Lineage class above
@classmethod
def add_to_tree(cls, lineage: Lineage):
if lineage.parent is None:
# No children to add
cls.tree.update(
{
'id': lineage.partial,
'data': lineage,
'parent': None,
}
)
elif 'children' in cls.tree:
count = len(cls.tree['children'].items())
cls.tree['children'][count] = {
'id': lineage.partial,
'data': lineage,
'parent': lineage.parent,
}
else:
cls.tree['children'] = {
0: {
'id': lineage.partial,
'data': lineage,
'parent': lineage.parent,
}
}
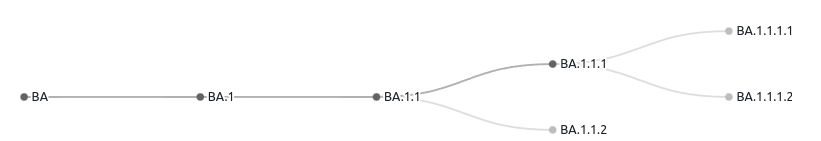
In the following dict object, you will see what the code produces. And below that is an image of the structure of the dataset.
As you can see, child index 1 should actually be a child of lineage at index 0. And Children indexes 2 and 5 should actually be children of lineage at index 1, and so on.
The problem is that everything is a child a the root. There is no further hierarchy. What are some ways to mitigate this issue?
{
"id": "BA",
"data": {
"designation_date": "2021-11-24",
"pango": "B.1.1.529",
"partial": "BA",
"unaliased": "B.1.1.529",
"parent": null
},
"parent": null,
"children": {
"0": {
"id": "BA.1",
"data": {
"designation_date": "2021-12-07",
"pango": "BA.1",
"partial": "BA.1",
"unaliased": "B.1.1.529.1",
"parent": "BA"
},
"parent": "BA"
},
"1": {
"id": "BA.1.1",
"data": {
"designation_date": "2022-01-07",
"pango": "BA.1.1",
"partial": "BA.1.1",
"unaliased": "B.1.1.529.1.1",
"parent": "BA.1"
},
"parent": "BA.1"
},
"2": {
"id": "BA.1.1.1",
"data": {
"designation_date": "2022-02-23",
"pango": "BA.1.1.1",
"partial": "BA.1.1.1",
"unaliased": "B.1.1.529.1.1.1",
"parent": "BA.1.1"
},
"parent": "BA.1.1"
},
"3": {
"id": "BA.1.1.1.1",
"data": {
"designation_date": "2022-02-23",
"pango": "BC.1",
"partial": "BA.1.1.1.1",
"unaliased": "B.1.1.529.1.1.1.1",
"parent": "BA.1.1.1"
},
"parent": "BA.1.1.1"
},
"4": {
"id": "BA.1.1.1.2",
"data": {
"designation_date": "2022-06-13",
"pango": "BC.2",
"partial": "BA.1.1.1.2",
"unaliased": "B.1.1.529.1.1.1.2",
"parent": "BA.1.1.1"
},
"parent": "BA.1.1.1"
},
"5": {
"id": "BA.1.1.2",
"data": {
"designation_date": "2022-02-23",
"pango": "BA.1.1.2",
"partial": "BA.1.1.2",
"unaliased": "B.1.1.529.1.1.2",
"parent": "BA.1.1"
},
"parent": "BA.1.1"
},
Again, this is how the data SHOULD be if it was in the correct heirachy. However, it isn't
Data
I discard the Github issues for now
"PANGO Lineage" "Partial-aliased PANGO Lineage" "Unaliased PANGO Lineage" "Designation Date" "GitHub Issue"
B.1.1.529 BA B.1.1.529 2021-11-24 #343
BA.1 BA.1 B.1.1.529.1 2021-12-07 #361
BA.1.1 BA.1.1 B.1.1.529.1.1 2022-01-07 #360
BA.1.1.1 BA.1.1.1 B.1.1.529.1.1.1 2022-02-23
BC.1 BA.1.1.1.1 B.1.1.529.1.1.1.1 2022-02-23
BC.2 BA.1.1.1.2 B.1.1.529.1.1.1.2 2022-06-13 #570
BA.1.1.2 BA.1.1.2 B.1.1.529.1.1.2 2022-02-23
CodePudding user response:
I would suggest adding a class attribute refs so you can keep a reference to all nodes in your tree by their id. That way you can quickly look up the parent of the node that must be added, and establish the parent-child link with it:
class Lineage:
tree = dict()
refs = dict()
# ...
@classmethod
def add_to_tree(cls, lineage: 'Lineage'):
node = {
"id": lineage.partial,
"lineage": lineage,
"parent": None,
"children": []
}
cls.refs[lineage.partial] = node
if lineage.parent is None:
cls.tree.update(node)
elif lineage.parent in cls.refs:
cls.refs[lineage.parent]["children"].append(node)
node["parent"] = cls.refs[lineage.parent]
This way of working requires that parents are added to the tree before their children. After loading all nodes into the tree you can empty or delete the refs dictionary, as it has served its purpose.
