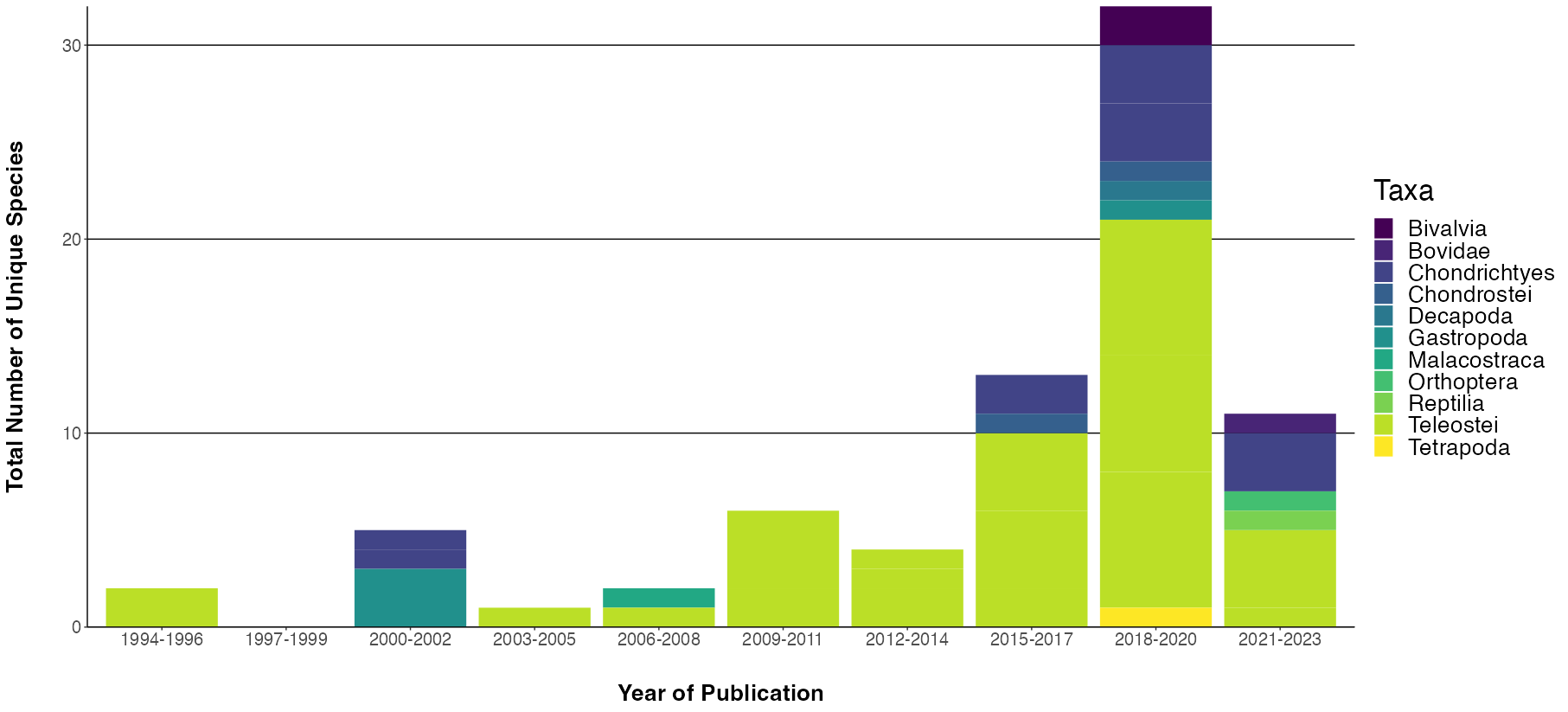
I am trying to plot some data that is assoicated with a year, and the years are not consecutive (e.g. some data was recorded in 1984, 1999, 2000, 2001 etc.). I want to bin my data into groups of 3 years (e.g. 1984-1986, 1987-1989 etc.), but I also want the binned years that have no data to show on the x axis.... so if there is no data for 1987-1989 I still want that on the x axis to show there was no data.
Here is my dataframe
taxa_diversity = structure(list(Year_Publication = c(1994L, 2001L, 2002L, 2002L,
2004L, 2006L, 2007L, 2009L, 2011L, 2012L, 2013L, 2014L, 2015L,
2016L, 2017L, 2017L, 2017L, 2018L, 2018L, 2018L, 2019L, 2019L,
2019L, 2020L, 2020L, 2020L, 2020L, 2021L, 2021L, 2022L, 2022L,
2022L, 2022L), Taxa = c("Teleostei", "Chondrichtyes", "Chondrichtyes",
"Gastropoda", "Teleostei", "Malacostraca", "Teleostei", "Teleostei",
"Teleostei", "Teleostei", "Teleostei", "Teleostei", "Teleostei",
"Teleostei", "Chondrichtyes", "Teleostei", "Chondrostei", "Teleostei",
"Chondrichtyes", "Decapoda", "Teleostei", "Gastropoda", "Chondrichtyes",
"Chondrostei", "Teleostei", "Bivalvia", "Tetrapoda", "Teleostei",
"Orthoptera", "Chondrichtyes", "Teleostei", "Reptilia", "Bovidae"
), Total_Species_Per_Taxa = c(2L, 1L, 1L, 3L, 1L, 1L, 1L, 2L,
4L, 2L, 1L, 1L, 2L, 4L, 2L, 4L, 1L, 7L, 3L, 1L, 6L, 1L, 3L, 1L,
7L, 2L, 1L, 1L, 1L, 3L, 4L, 1L, 1L), Total_Species_Per_Pub = c(2L,
1L, 4L, 4L, 1L, 1L, 1L, 2L, 4L, 2L, 1L, 1L, 2L, 4L, 7L, 7L, 7L,
11L, 11L, 11L, 10L, 10L, 10L, 11L, 11L, 11L, 11L, 2L, 2L, 9L,
9L, 9L, 9L)), class = "data.frame", row.names = c(NA, -33L))
This is code I'm using the make the graph I currently have
Taxa_Plot = ggplot(data = taxa_diversity,
aes(x = factor(`Year_Publication`),
y = `Total_Species_Per_Taxa`, fill = Taxa))
geom_col()
scale_fill_viridis(discrete = TRUE)
theme_bw()
theme(
#get rid of grey grid marks in the back
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.grid.major.x = element_blank() ,
# set horizontal grid marks to follow y-values across
panel.grid.major.y = element_line( size= .5, color="black" ),
panel.background = element_blank(),
panel.border = element_blank(),
axis.line = element_line(colour = "black"),
axis.title.x = element_text(size=20, face="bold",
margin = margin(30,0,0,0)),
axis.title.y = element_text(size=20, face="bold",
margin = margin(0,30,0,0)),
#increase the text size on x and y axis
axis.text.x = element_text(size = 15),
axis.text.y = element_text(size = 15),
legend.text = element_text(size = 20),
legend.title = element_text(size = 25))
#change axis titles
xlab("Year of Publication")
ylab("Total Number of Unique Species")
scale_y_continuous(limits = c(0,14),
expand = expansion(mult = c(0,0)))
Taxa_Plot
I was hoping there might be a way to call for the binning in the ggplot functions....
Any ideas.
CodePudding user response:
Using cut to create your bins and setting drop=FALSE in scale_x_discrete you could do:
library(ggplot2)
library(viridis)
breaks <- seq(1994, 2024, by = 3)
labels <- paste(breaks[-length(breaks)], breaks[-1] - 1, sep = "-")
taxa_diversity$Year_Publication_Cut <- cut(
taxa_diversity$Year_Publication,
breaks, labels, right = FALSE)
ggplot(
data = taxa_diversity,
aes(
x = Year_Publication_Cut,
y = Total_Species_Per_Taxa,
fill = Taxa
)
)
geom_col()
scale_x_discrete(drop = FALSE)
scale_y_continuous(
#limits = c(0, 14),
expand = expansion(mult = c(0, 0))
)
scale_fill_viridis(discrete = TRUE)
theme_bw()
theme(
# get rid of grey grid marks in the back
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.grid.major.x = element_blank(),
# set horizontal grid marks to follow y-values across
panel.grid.major.y = element_line(size = .5, color = "black"),
panel.background = element_blank(),
panel.border = element_blank(),
axis.line = element_line(colour = "black"),
axis.title.x = element_text(
size = 20, face = "bold",
margin = margin(30, 0, 0, 0)
),
axis.title.y = element_text(
size = 20, face = "bold",
margin = margin(0, 30, 0, 0)
),
# increase the text size on x and y axis
axis.text.x = element_text(size = 15),
axis.text.y = element_text(size = 15),
legend.text = element_text(size = 20),
legend.title = element_text(size = 25)
)
# change axis titles
xlab("Year of Publication")
ylab("Total Number of Unique Species")
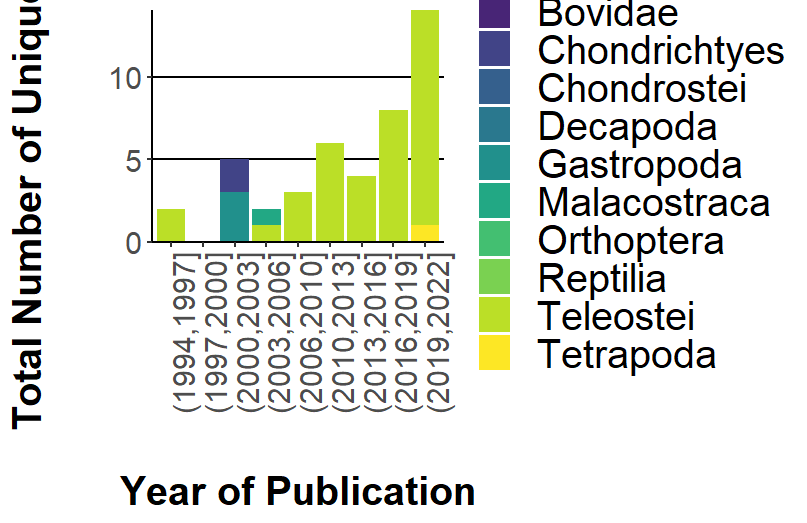
CodePudding user response:
You can transform your data this way:
taxa_diversity <- transform(taxa_diversity, Year_Publication =
cut(Year_Publication, (max(Year_Publication)-min(Year_Publication)) / 3))
add a discrete scale:
scale_x_discrete(drop=FALSE)
to clarity you set the angle of labels:
...
axis.text.x = element_text(size = 15, angle = 90),
...
The plot will be shown as follows:
CodePudding user response:
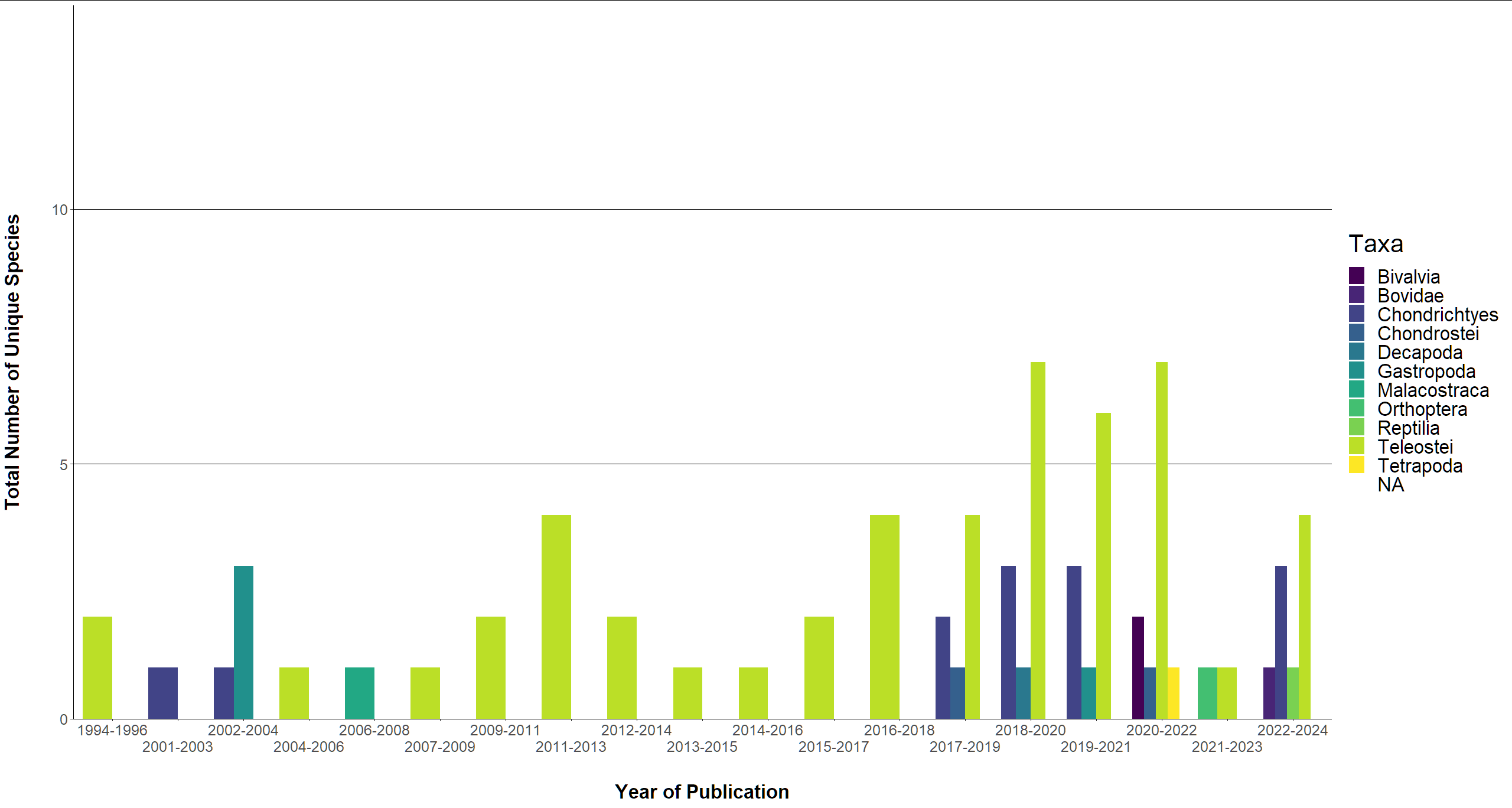
I am not sure, maybe this is also an option for you:
library(viridis)
library(tidyverse)
taxa_diversity %>%
arrange(Year_Publication) %>%
mutate(id = row_number()) %>%
group_by(group = cumsum(Year_Publication != lag(Year_Publication, def = first(Year_Publication)))) %>%
mutate(Year_Publication1 = Year_Publication 2) %>%
pivot_longer(c(Year_Publication, Year_Publication1)) %>%
arrange(name, .by_group = TRUE) %>%
tidyr::complete(value = seq(min(value), max(value))) %>%
mutate(labelx = paste0(min(value), "-", max(value))) %>%
#-------------------------------------------------------
ggplot(aes(x = factor(labelx),
y = `Total_Species_Per_Taxa`, fill = Taxa))
geom_col(position = position_dodge()) # added
scale_fill_viridis(discrete = TRUE)
theme_bw()
theme(
#get rid of grey grid marks in the back
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.grid.major.x = element_blank() ,
# set horizontal grid marks to follow y-values across
panel.grid.major.y = element_line( size= .5, color="black" ),
panel.background = element_blank(),
panel.border = element_blank(),
axis.line = element_line(colour = "black"),
axis.title.x = element_text(size=20, face="bold",
margin = margin(30,0,0,0)),
axis.title.y = element_text(size=20, face="bold",
margin = margin(0,30,0,0)),
#increase the text size on x and y axis
axis.text.x = element_text(size = 15),
axis.text.y = element_text(size = 15),
legend.text = element_text(size = 20),
legend.title = element_text(size = 25))
#change axis titles
xlab("Year of Publication")
ylab("Total Number of Unique Species")
scale_y_continuous(limits = c(0,14),
expand = expansion(mult = c(0,0)))
scale_x_discrete(guide = guide_axis(n.dodge = 2)) # added