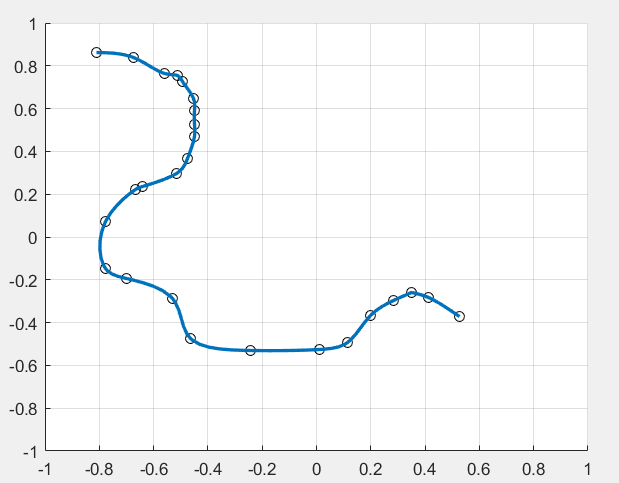
I am using MatLab to pick the data point with my mouse and then fit it with a spline. I found the following function could do the job
[xy, spcv] = getcurve()
it returns the x & y of the points I picked with a mouse
x = [ -0.8103 -0.6740 -0.5599 -0.5120 -0.4936 -0.4530 -0.4494 -0.4494 -0.4494 -0.4751 -0.5157 -0.6409 -0.6667 -0.7772 -0.7772 -0.6998 -0.5304 -0.4641 -0.2431 0.0110 0.1142 0.1989 0.2836 0.3499 0.3499 0.4125 0.5267]
y = [0.8621 0.8388 0.7640 0.7547 0.7266 0.6472 0.5911 0.5257 0.4696 0.3668 0.2967 0.2360 0.2220 0.0724 -0.1472 -0.1939 -0.2874 -0.4743 -0.5304 -0.5257 -0.4930 -0.3668 -0.2967 -0.2593 -0.2593 -0.2827 -0.3715
plotting the spline spcv returned by getcurve(), I get the following figure
By reading the code of getcurve, I see that it uses cscvn to return a parametric `natural' cubic spline that interpolates to the given points. It may be the reason why the curve is passing all given points instead of the best-fit curve. I would like to replace it with a cubic smoothing spline as below
sp = spaps(x, y, 0.);
xx = -1:0.01:2;
plot(xx, fnval(sp, xx), 'b', 'linewidth', 2); hold on;
plot(x, y, 'ko');
which gives me something strange as follow
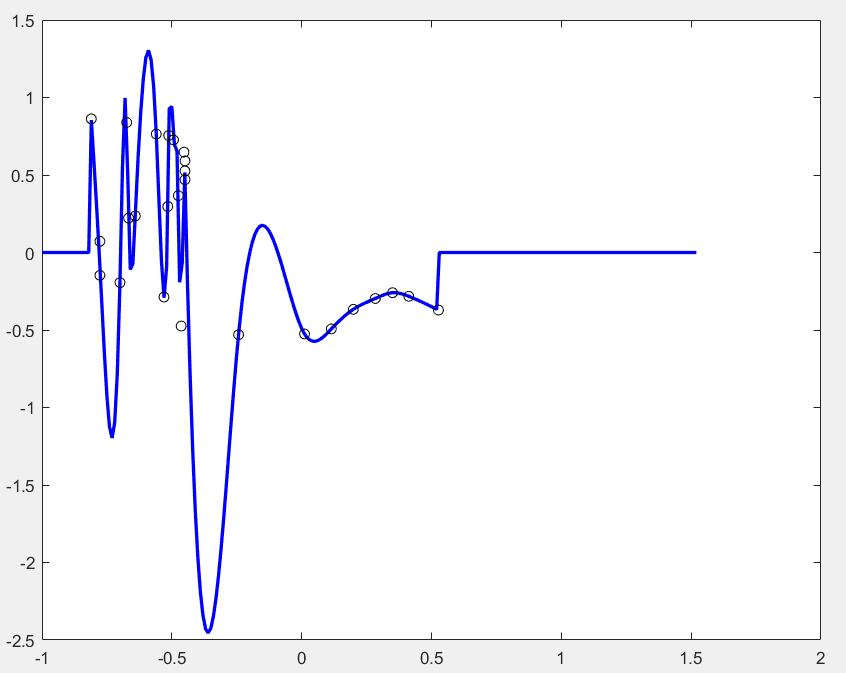
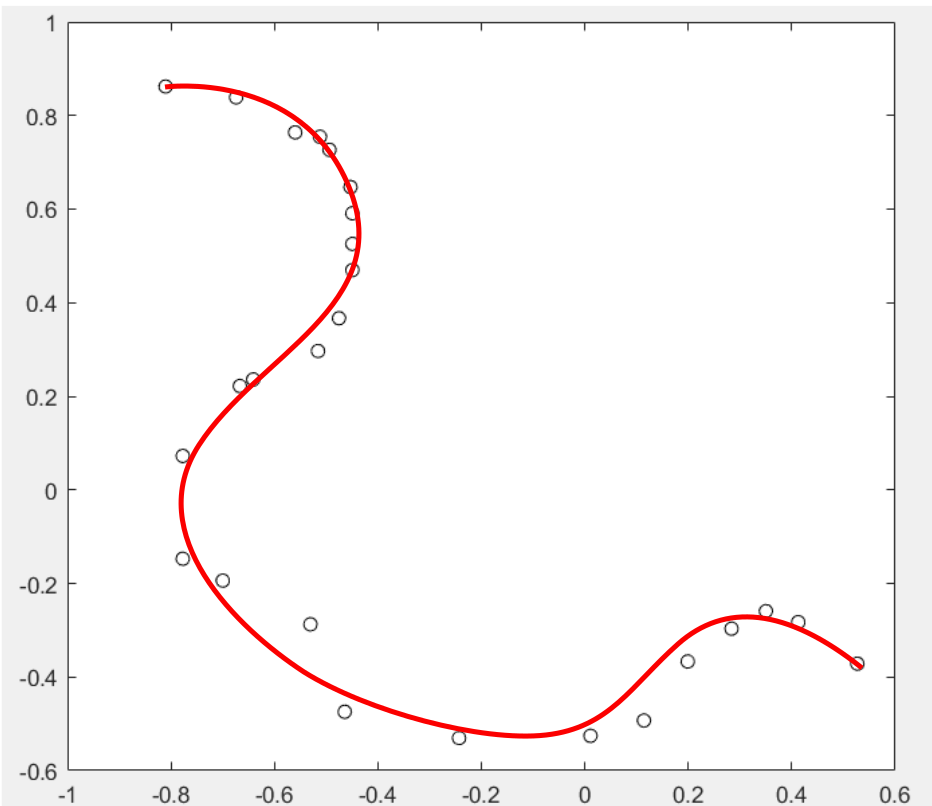
It looks like it is trying to fit the data points from small x to big x instead of the order given by the data sequence. I am expecting a smooth curve in red as below (I draw it by hand)
I am looking for a solution that I can replace cscvn with spaps in getcurve and keep the correct order. Thanks.
CodePudding user response:
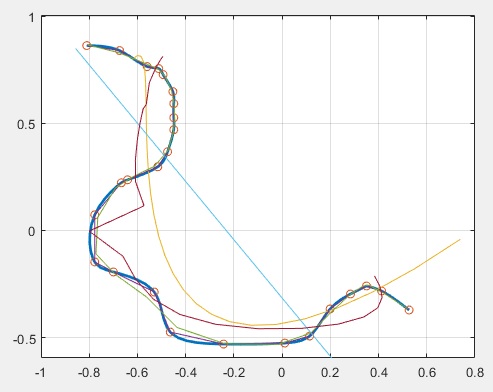
1.- To interpolate curves that fold back you have to parametrize x and y
x = [ -0.8103 -0.6740 -0.5599 -0.5120 -0.4936 -0.4530 -0.4494 -0.4494 -0.4494 -0.4751 -0.5157 -0.6409 -0.6667 -0.7772 -0.7772 -0.6998 -0.5304 -0.4641 -0.2431 0.0110 0.1142 0.1989 0.2836 0.3499 0.3499 0.4125 0.5267];
y = [0.8621 0.8388 0.7640 0.7547 0.7266 0.6472 0.5911 0.5257 0.4696 0.3668 0.2967 0.2360 0.2220 0.0724 -0.1472 -0.1939 -0.2874 -0.4743 -0.5304 -0.5257 -0.4930 -0.3668 -0.2967 -0.2593 -0.2593 -0.2827 -0.3715];
figure(1)
ax1=gca
fnplt(cscvn([x;y])); hold(ax1,'on');
plot(ax1,x,y,'o');
grid on
sp = spaps(x, y, 0.);
xx = -1:0.01:2;
figure(2)
plot(xx, fnval(sp, xx), 'b', 'linewidth', 2); hold on;
plot(x, y, 'ko'); grid on
nx=[1:numel(x)];ny=[1:numel(y)];
fx=fit(nx',x','poly3','Normalize','on','Robust','Bisquare')
fy=fit(ny',y','poly3','Normalize','on','Robust','Bisquare')
plot(ax1,fx(nx),fy(ny))
f2x=fit(nx',x','cubicinterp','Normalize','on')
f2y=fit(ny',y','cubicinterp','Normalize','on')
plot(ax1,f2x(nx),f2y(ny))
f3x=fit(nx',x','smoothingspline','Normalize','on')
f3y=fit(ny',y','smoothingspline','Normalize','on')
plot(ax1,f3x(nx),f3y(ny))
f4x=fit(nx',x','poly1','Normalize','on')
f4y=fit(ny',y','poly1','Normalize','on')
plot(ax1,f4x(nx),f4y(ny))
f5x=fit(nx',x','poly11','Normalize','on')
f5y=fit(ny',y','poly11','Normalize','on')
plot(ax1,f5x(nx),f5y(ny))
f6x=fit(nx',x','rat33','Normalize','on')
f6y=fit(ny',y','rat33','Normalize','on')
plot(ax1,f6x(nx),f6y(ny))
2.- Note that nx and ny have same amount of samples as x and y.
If you interpolate x over nx and y over ny and then apply fit the resulting curve will get closer to all points in the way you asked for.