I'm trying to write a Python function that takes an RGB image, looks at each pixel's RGB values (0-255) and simply finds the mean (average) of the three values. I wrote the following function:
def avgcolors(img_matrix):
M, N, D = img_matrix.shape
new_img = np.zeros((M, N))
for i in range(M):
for j in range(N):
pixel_mean = int(np.round(np.mean(img_matrix[i][j])))
new_img[i][j] = pixel_mean
return(new_img)
To me the problem seems very straight forward. Run through the image matrix indexing pixels with i, j. Find the mean of the three values in each pixel and write that to a new image matrix. However, something strange happens when I run the following:
pic = Image.open("red.jpg")
img = np.array(pic.getdata()).reshape(pic.size[0], pic.size[1], 3)
grayscale = avgcolors(img)
plt.imshow(grayscale, cmap='gray')
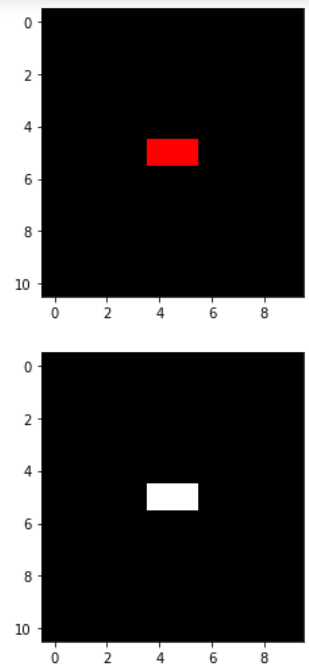
The "red.jpg" image and the output are shown below:
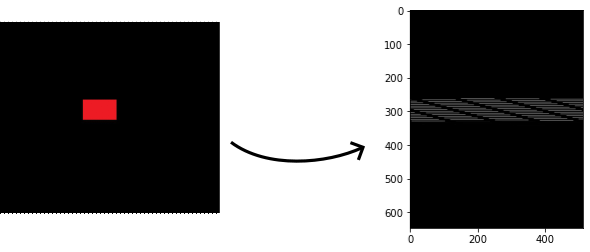
I would have thought that when the script reaches the red pixels it would average out the values 255, 0, 0 to get a grayish pixel with a value of 85. And elsewhere, it would just be black. Instead, I am not getting that and I get strange diagonal artifacts. What am I doing wrong? I can't figure it out.
CodePudding user response:
I've found the problem. The problem was in converting the image to a numpy array. It seems the command
img = np.array(pic.getdata()).reshape(pic.size[0], pic.size[1], 3)
Rearranges the array in a strange way. I've simply changed this line of code for:
img = np.array(pic)
CodePudding user response:
Here is an implementation of this using lists.
image = np.array([
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*4,[[255,0,0]]*2,[[0,0,0]]*4]),
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*10]),
np.concatenate([[[0,0,0]]*10])
])
image = image.tolist()
def mean_image_explicit(image):
rows = []
for i in range(len(image)):
cols = []
for j in range(len(image[0])):
v = sum(image[i][j]) / len(image[i][j])
cols.append(v)
rows.append(cols)
return rows
def mean_image(image):
rows = [[sum(image[i][j]) / len(image[i][j]) for j in range(len(image[0]))] for i in range(len(image))]
return rows
avg = mean_image(image)
plt.imshow(image)
plt.show()
plt.imshow(avg,cmap='gray')
plt.show()
CodePudding user response:
I hope I'm understanding correctly... if you are trying to compute the mean of an 3-dimensional array on a specific axis, you can use np.array.mean(axis=2):
import numpy as np
import matplotlib.pyplot as plt
image = np.random.randint(0, 255, size=(10, 10, 3), dtype=np.uint8)
print(image.shape) # (10,10,3)
plt.imshow(image)
plt.show()
mean_rgb = image.mean(axis=2)
print(mean_rgb.shape) # (10,10)
plt.imshow(mean_rgb,cmap="Greys")
plt.show()
CodePudding user response:
You can simply use img = np.array(pic) because PIL and numpy understand each other like that.
The .getdata() call should not be used. It is less efficient.
The .reshape did garble the data, but doesn't necessarily.
The issue you're having there is that you mixed up width and height.
PIL.Image.sizeis a(width, height)tuple.- Numpy requires the shape to be
(height, width, channels).
If you use the shape (pic.size[1], pic.size[0], 3), your original code would work just fine.