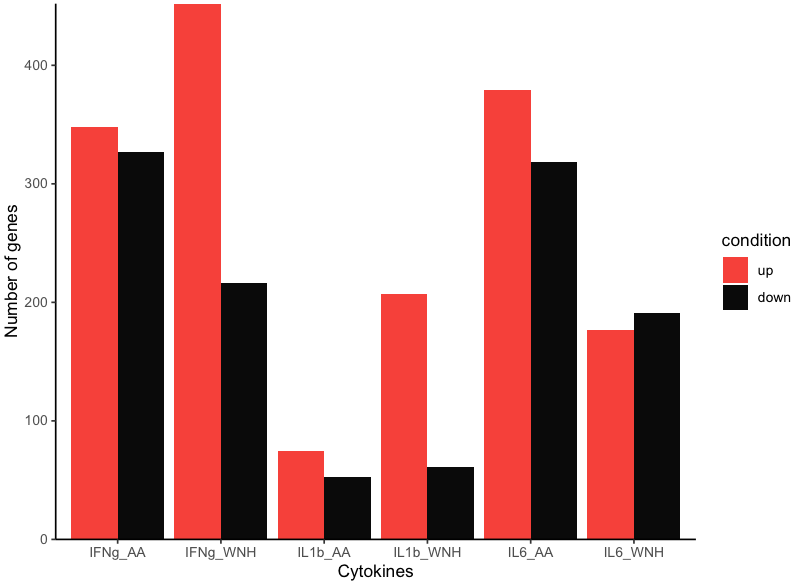
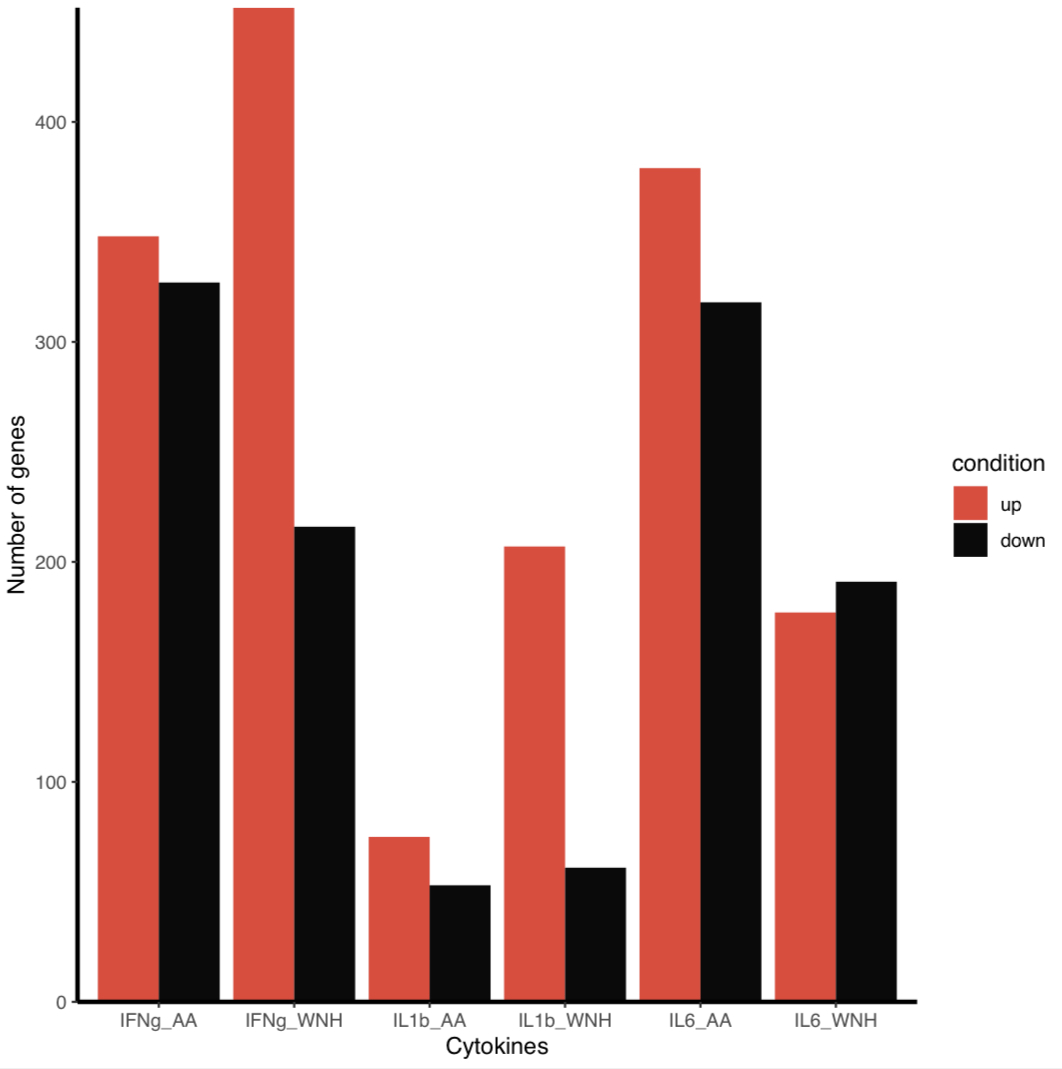
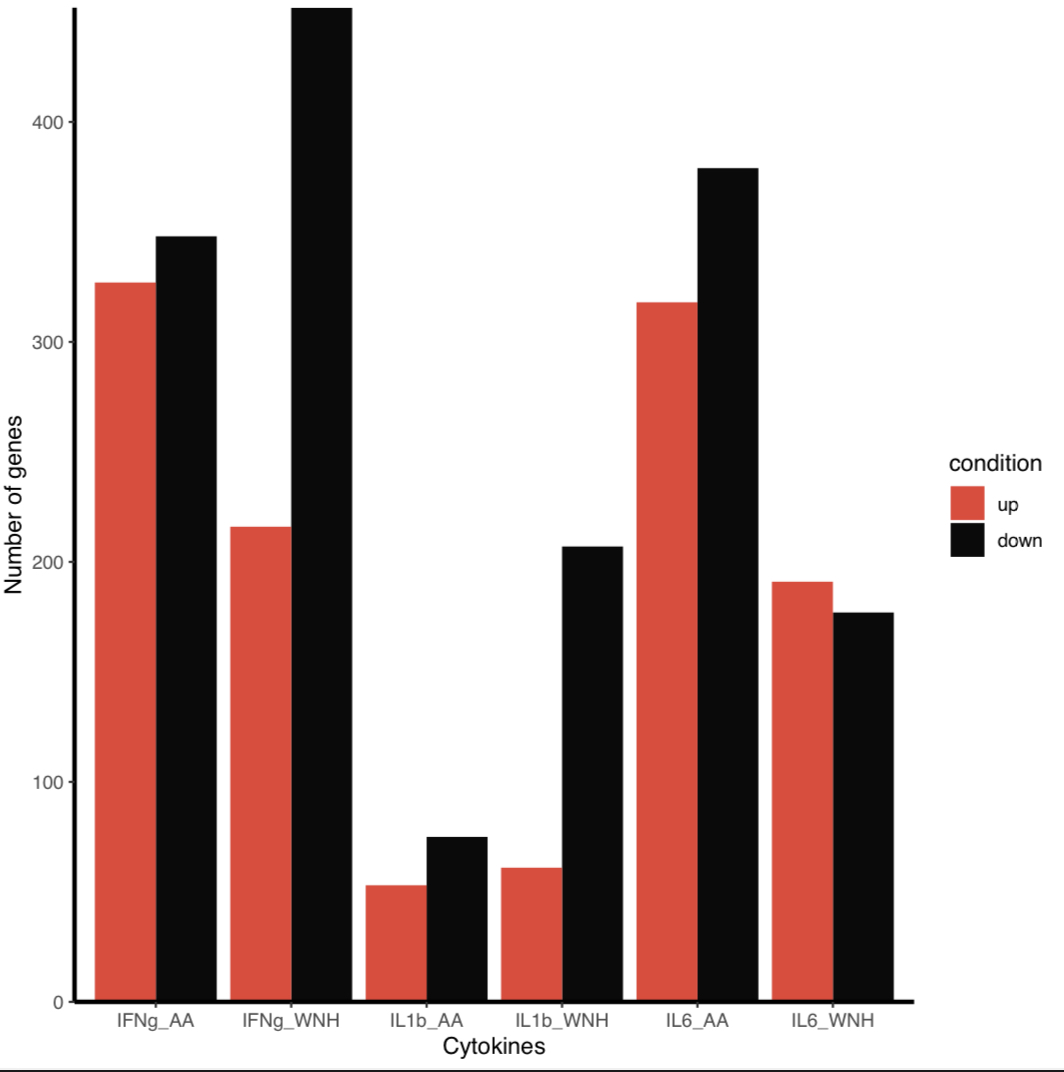
I want my bars to correspond to a legend and to have them colored with my own color, not the default one.
# library
library(ggplot2)
# create a dataset
specie <- c(rep("IFNg_WNH", 2) ,
rep("IFNg_AA", 2),
rep("IL1b_WNH", 2),
rep("IL1b_AA", 2),
rep("IL6_WNH", 2),
rep("IL6_AA", 2)
)
condition <- rep(c("down", "up"), 6)
value <- c(452,216,
348,327,
207,61,
75,53,
177,191,
379,318)
data <- data.frame(specie,condition,value)
data
# Grouped
p <- ggplot(data, aes(fill=condition, y=value, x=specie))
geom_bar(position="dodge", stat="identity")
z <- p labs(y = "Number of genes", x = "Cytokines")
theme_classic()
theme(plot.title = element_text(hjust = 0.5))
theme(axis.line=element_line(size=1))
scale_y_continuous(expand = c(0, 0), limits = c(0, NA))
scale_fill_discrete(labels=c('up', 'down'))
z
Once I add
z scale_fill_manual(values=c('#eb4034','#0a0a0a'))
The color is changing but the legend reverts to the wrong one. What is happening?
CodePudding user response:
Adding type to scale_fill_discrete
type: One of the following:
• A character vector of color codes.
• A list of character vectors of color codes.
• A function that returns a discrete colour/fill scale
ggplot(data, aes(fill=condition, y=value, x=specie))
geom_bar(position="dodge", stat="identity")
labs(y = "Number of genes", x = "Cytokines")
theme_classic()
theme(plot.title = element_text(hjust = 0.5))
scale_y_continuous(expand = c(0, 0), limits = c(0, NA))
scale_fill_discrete(labels=c('up', 'down'), type=c('#eb4034','#0a0a0a'))
CodePudding user response:
There are a couple issues here. First, scale_fill_manual() essentially “overwrites” scale_fill_discrete(). Instead, use just one scale_fill_*() call including all relevant arguments:
library(ggplot2)
p
labs(y = "Number of genes", x = "Cytokines")
theme_classic()
theme(plot.title = element_text(hjust = 0.5))
theme(axis.line=element_line(size=1))
scale_y_continuous(expand = c(0, 0), limits = c(0, NA))
scale_fill_manual(labels = c('up', 'down'), values=c('#eb4034', '#0a0a0a'))
However - currently, the labels argument is in effect recoding your data, so that "up" values are labeled "down" and vice versa, which I assume isn’t what you want. My best guess is you’re actually trying to change the order the labels appear in the legend. If so, you can change the factor levels of condition to the order you want:
data$condition <- factor(data$condition, c("up", "down"))
p <- ggplot(data, aes(fill=condition, y=value, x=specie))
geom_bar(position="dodge", stat="identity")
p
labs(y = "Number of genes", x = "Cytokines")
theme_classic()
theme(plot.title = element_text(hjust = 0.5))
theme(axis.line=element_line(size=1))
scale_y_continuous(expand = c(0, 0), limits = c(0, NA))
scale_fill_manual(values=c('#eb4034', '#0a0a0a'))