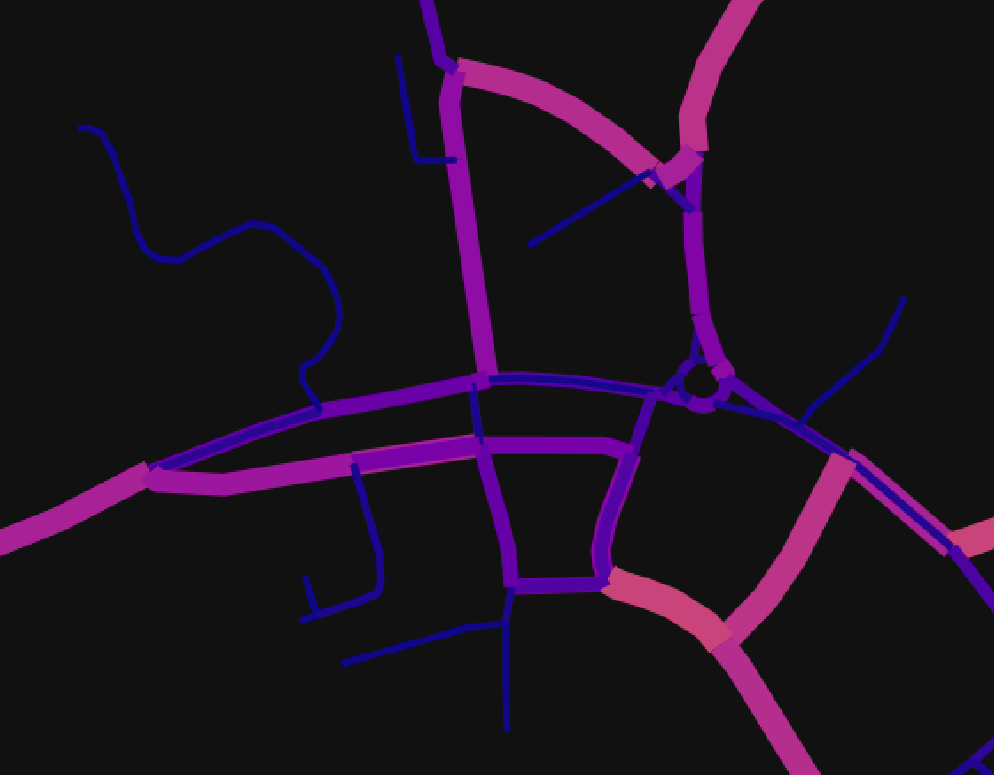
I'm using OSMnx to create some plots of road networks with data shown by colour, below is an example which produces an image with colour showing betweenness centrality. The problem I'm having is that, due to the zordering, some of the lighter regions are covered by other edges, especially around junctions with lots of nodes.
The OSMnx.plot_graph method calls gdf.plot from GeoPandas, which in turn uses PlotAccessor from Pandas. I've been reading the OSMnx documentation, and can't seem to find a way to pass a z-ordering, so my question is: is there a way I can directly plot the graph, either with GeoPandas, pandas, or even matplotlib directly, such that I can specify a z-ordering of the edges?
import networkx as nx
import osmnx as ox
place = 'Hornsea'
G = ox.graph_from_place(place, network_type="drive")
# Digraph removes parallel edges
# Line graph swaps nodes and edges
line_graph = nx.line_graph(ox.get_digraph(G))
betweenness_centrality = nx.betweenness_centrality(line_graph, weight='travel_time')
for edge in G.edges:
if edge not in betweenness_centrality:
nx.set_edge_attributes(G, {edge: 0}, 'betweenness_centrality')
betweenness_centrality = {(k[0], k[1], 0): v for k, v in betweenness_centrality.items()}
nx.set_edge_attributes(G, betweenness_centrality, "betweenness_centrality")
ec = ox.plot.get_edge_colors_by_attr(G, 'betweenness_centrality', cmap='plasma') # "RdYlGn_r"
ew = [G.get_edge_data(*edge).get('betweenness_centrality', 0) * 10 0.25 for edge in G.edges]
fig, ax = ox.plot_graph(G, edge_color=ec, edge_linewidth=ew, node_size=0)
fig.savefig(f"images/{place}_betweenness_centrality.png", facecolor="w", dpi=1000, bbox_inches="tight")
CodePudding user response:
Took a little exploring the source code... not sure if this is exactly right, since I'm not sure how the desired outcome is supposed to look, but hopefully this gets you most of the way there... had to do some poking in the source code....
from pathlib import Path
import geopandas as gpd
import matplotlib.pyplot as plt
import networkx as nx
import numpy as np
import osmnx as ox
from shapely.geometry import LineString
from shapely.geometry import Point
# From your example:
place = 'Hornsea'
G = ox.graph_from_place(place, network_type='drive')
line_graph = nx.line_graph(ox.get_digraph(G))
betweenness_centrality = nx.betweenness_centrality(
line_graph, weight='travel_time')
for edge in G.edges:
if edge not in betweenness_centrality:
nx.set_edge_attributes(G, {edge: 0}, 'betweenness_centrality')
betweenness_centrality = {
(k[0], k[1], 0): v for k, v in betweenness_centrality.items()}
nx.set_edge_attributes(
G, betweenness_centrality, 'betweenness_centrality')
ec = ox.plot.get_edge_colors_by_attr(
G, 'betweenness_centrality', cmap='plasma')
CodePudding user response:
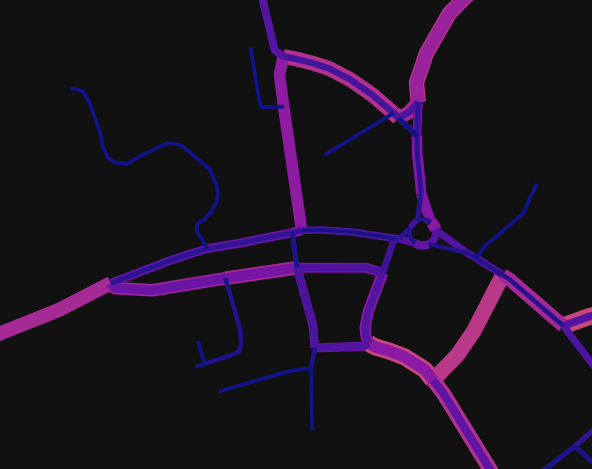
Thanks to Damian Satterthwaite-Phillips, I was able to find a solution to the problem. His answer can be cut down a lot, so I thought I'd share what I ended up using in case anyone else has the same issue. However, I have made a pull request to have edge_zorder be an argument for osmnx.plot_graph, so hopefully soon this won't be necessary.
def plot_graph(G, edge_colour=None, edge_linewidth=None, edge_zorder=None, edge_alpha=None, show=True, save=False, close=False, filepath=None, dpi=300):
fig, ax = plt.subplots(figsize=(8, 8), facecolor='#111111', frameon=False)
ax.set_facecolor('#111111')
gs_edges = ox.utils_graph.graph_to_gdfs(G, nodes=False)["geometry"]
gdf_edges = gpd.GeoDataFrame(gs_edges)
gdf_edges['zorder'] = edge_zorder
gdf_edges['colour'] = edge_colour
gdf_edges['linewidth'] = edge_linewidth
gdf_edges['alpha'] = edge_alpha
gdf_edges.sort_values('zorder', inplace=True)
colour = gdf_edges.colour if edge_colour is not None else None
linewidth = gdf_edges.linewidth if edge_linewidth is not None else None
alpha = gdf_edges.alpha if edge_alpha is not None else None
ax = gdf_edges.plot(ax=ax, color=colour, lw=linewidth, alpha=alpha, zorder=1)
west, south, east, north = gdf_edges.total_bounds
bbox = north, south, east, west
ax = ox.plot._config_ax(ax, G.graph["crs"], bbox, 0.02)
return ox.plot._save_and_show(fig, ax, save, show, close, filepath, dpi)
Now this doesn't plot nodes, but it would be easily modified by looking at the plot_graph method in osmnx.plot.py