I am trying to make some violin plots with ggplot using this dataframe
df = structure(list(nid.weight = c(2.46, 0.319, 1.169, 1.631, 2.03,
0.148, 0.252, 5.614, 2.557, 6.062, 2.939, 6.04, 18.858, 28.727,
18.3, 9.831, 16.298, 17.176, 13.391, 15.044, 35.42, 5.421, 10.073,
15.499, 12.712, 16.046, 23.003, 11.656, 19.79, 20.593, 19.264,
26.35, 13.752, 31.795, 18.604, 18.871, 22.848, 34.46, 14.176,
20.73, 31.97, 18.7, 17.837, 15.875, 14.44, 38.78, 14.595, 21.522,
13.041, 18.051, 20.748, 17.91, 14.831, 9.523, 9.865, 38.2, 19.531,
25.724, 16.208, 18.059, 14.98, 11.9, 14.9, 13, 16.3, 15.555,
0.031, 1.99, 8.924, 21.081, 30.12, 8.658, 0.078, 0.111, 0.373,
0.217, 0.276, 20.993, 12.936, 0.142, 0.188, 0.154, 0.182, 0.14,
0.172, 0.123, 0.187, 0.104, 17.903, 0.18, 21.026, 0.124, 0.108,
21.394, 0.14, 0.189, 0.173, 0.271, 0.124, 0.122, 0.097, 0.16,
8.087, 0.107, 0.149, 0.072, 16.732, 12.663, 0.268, 0.268, 0.315,
0.277, 0.154, 0.233, 0.323, 14.043, 0.424, 0.296, 0.531, 0.287,
0.45, 0.248, 0.475, 0.726, 0.379, 0.623, 0.257, 0.558, 0.34,
13.687, 0.722, 14.936, 0.243, 0.731, 0.216, 0.4, 0.476, 0.114,
1.46, 0.861, 0.861, 1.64, 0.655, 1.096, 0.649, 0.789, 1.083,
1.072, 1.748, 1.404, 0.721, 1.026, 0.305, 0.59, 0.929, 0.937,
1.984, 1.078, 1.632, 3.373, 2.183, 0.546, 2.745, 2.598, 0.789,
0.925, 0.636, 1.184, 1.171, 1, 1.229, 1.503, 1.172, 1.89, 0.946,
0.641, 0.701, 0.228, 0.169, 0.389, 0.894, 3.299, 1.491, 3.022,
1.395, 1.472, 0.7, 1.195, 0.865, 2.414, 0.442, 1.282, 1.228,
1.403, 0.655, 1.34, 2.014, 1.612, 1.08, 0.326, 1.131, 1.133,
1.362, 2.424, 0.565, 0.67, 1.04, 0.997, 1.022, 0.48, 0.837, 0.746,
0.483, 0.696, 0.934, 1.105, 0.86, 0.75, 0.82, 0.48, 2.437, 0.372,
0.234, 0.099, 0.051, 2.716, 0.621, 0.611, 0.384, 0.82, 0.646,
0.68, 0.768, 0.378, 0.305, 2.462, 2.185, 0.598, 1.529, 2.175,
5.242, 7.084, 0.105, 1.29, 1.154, 2.961, 6.741, 1.742, 1.632,
3.47, 1.232, 2.359, 0.111, 1.638, 2.38, 1.162, 5.291, 1.114,
0.487, 0.874, 0.564, 1.318, 5.55, 7.685, 2.543, 0.401, 6.578,
7.53, 7.89, 3.312, 2.555, 0.233, 7.749, 1.289, 0.94, 0.839, 3.408,
6.603, 10.832, 7.353, 8.789, 5.352, 8.341, 3.897, 21.308, 16.963,
14.393, 3.852, 26.156, 21.705, 8.573, 9.504, 8.813, 2.458, 2.22,
32.4, 10.468, 7.66, 18.072, 2.135, 20.67, 4.79, 15.467, 8.484,
4.28, 13.36, 3.515, 7.835, 9.168, 2.443, 4.076, 9.953, 3.515,
5.206, 11.493, 3.059, 5.311, 7.07, 0.045, 5.309, 0.52, 9.56,
19.989, 36.894, 30.305, 21.25, 20.387, 10.685, 26.185, 0.404,
25.427, 5.755, 16.112, 14.832, 16.072, 14.835, 7.67, 8.717, 17.025,
19.564, 30.922, 0.049, 0.632, 0.415, 6.621, 13.701, 21.269, 17.527,
18.9, 16.574, 22.877, 28.866, 27.756, 7.535, 13.557, 19.082,
8.287, 18.617, 17.219, 14.733, 14.484, 12.481, 6.201, 35.361,
19.888, 24.468, 19.198, 29.679, 22.218, 29.408, 36.102, 23.984,
13.494, 30.313, 18.847, 0.731, 6.166, 28.418, 17.481, 20.235,
31.187, 26.49, 32.56, 14.459, 15.121, 2.385, 31.06, 14.626, 18.43,
9.808, 10.926, 10.1, 18.711, 26.396, 17.722, 12.006, 8.995, 17.874,
15.124, 10.318, 15.23, 22.661, 11.005, 6.016, 22.408, 7.561,
13.97, 8.252, 14.08, 10.254, 15.43, 25.756, 14.52, 9.588, 8.775,
29.909, 24.27, 10.459, 18.974, 11.11, 20.189, 16.73, 14.201,
28.025, 19.849, 20.307, 24.715, 10.688, 13.465, 15.817, 21.798,
19.616, 18.622, 12.703, 15.037, 24.377, 21.071, 10.81, 16.02,
15.576, 36.77, 21.363, 17.874, 19.724, 14.749, 9.152, 16.923,
0.065, 37.676, 25.147, 19.729, 18.345, 14.74, 14.938, 16.49,
20.211, 11.397, 15.34, 11.787, 12.373, 11.504, 10.563, 13.459,
12.091, 14.487, 7.769, 10.006, 9.041, 8.031, 9.05, 1.856, 3.405,
0.036, 12.772, 12.104, 8.282, 10.581, 4.867, 11.029, 10.558,
11.115, 16.303, 11.409, 12.732, 11.417, 11.352, 16.167, 23.197,
15.232, 17.714, 14.234, 23.325, 13.902, 13.66, 17.23, 15.176,
20.037, 15.751, 25.133, 15.217, 29.949, 24.001, 26.291, 39.325,
0.101, 0.148, 0.095, 0.194, 0.112, 0.07, 0.13, 0.096, 0.151,
15.518, 11.961, 19.033, 10.798, 0.114, 17.396), Fmaturity = structure(c(3L,
1L, 2L, 2L, 3L, 1L, 2L, 3L, 3L, 4L, 3L, 4L, 4L, 5L, 4L, 4L, 4L,
5L, 4L, 5L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 4L, 5L, 4L, 5L, 4L, 5L,
5L, 5L, 5L, 4L, 5L, 5L, 5L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 4L, 5L, 4L, 4L, 4L, 4L, 4L, 5L, 4L, 4L, 4L, 4L, 4L, 5L, 5L,
5L, 1L, 2L, 5L, 4L, 5L, 4L, 1L, 1L, 2L, 2L, 2L, 5L, 5L, 2L, 1L,
2L, 2L, 1L, 1L, 1L, 2L, 2L, 5L, 2L, 5L, 2L, 2L, 5L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 1L, 4L, 2L, 2L, 2L, 4L, 5L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 4L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 2L, 2L, 3L, 2L, 2L, 2L,
4L, 3L, 4L, 2L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
1L, 2L, 2L, 2L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L,
3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L,
3L, 1L, 2L, 2L, 2L, 3L, 3L, 2L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 3L,
2L, 2L, 2L, 2L, 2L, 3L, 3L, 2L, 2L, 3L, 3L, 3L, 2L, 2L, 2L, 3L,
3L, 2L, 2L, 2L, 3L, 4L, 3L, 3L, 3L, 3L, 3L, 5L, 3L, 3L, 3L, 4L,
4L, 3L, 4L, 4L, 2L, 2L, 4L, 5L, 3L, 5L, 2L, 4L, 2L, 4L, 3L, 3L,
4L, 3L, 3L, 3L, 3L, 2L, 4L, 2L, 3L, 4L, 3L, 3L, 3L, 1L, 3L, 2L,
5L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 2L, 5L, 3L, 4L, 4L, 4L, 4L, 4L,
5L, 5L, 4L, 4L, 1L, 2L, 2L, 3L, 4L, 5L, 5L, 4L, 5L, 4L, 5L, 4L,
4L, 5L, 5L, 3L, 4L, 4L, 5L, 4L, 5L, 4L, 5L, 5L, 5L, 4L, 5L, 4L,
4L, 4L, 5L, 4L, 5L, 4L, 2L, 3L, 5L, 5L, 5L, 5L, 4L, 5L, 4L, 5L,
2L, 5L, 4L, 4L, 4L, 5L, 4L, 5L, 5L, 5L, 5L, 4L, 5L, 5L, 5L, 5L,
5L, 5L, 4L, 5L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 4L, 5L, 5L, 5L,
5L, 5L, 4L, 5L, 5L, 5L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
4L, 4L, 5L, 5L, 5L, 5L, 4L, 5L, 5L, 4L, 4L, 5L, 5L, 5L, 1L, 4L,
4L, 5L, 4L, 4L, 5L, 5L, 5L, 5L, 4L, 5L, 5L, 5L, 4L, 5L, 5L, 5L,
4L, 4L, 4L, 5L, 4L, 2L, 3L, 1L, 4L, 4L, 5L, 5L, 3L, 5L, 5L, 5L,
5L, 5L, 4L, 5L, 5L, 5L, 5L, 5L, 4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 4L, 5L, 4L, 4L, 1L, 2L, 2L, 2L, 2L, 1L, 2L, 1L, 1L,
5L, 5L, 5L, 4L, 2L, 4L), levels = c("1", "2", "3", "4", "5"), class = "factor")), class = "data.frame", row.names = c(NA,
-519L))
Starting out I tried using the simplest code
ggplot(df, aes(x=Fmaturity, y=nid.weight))
geom_violin()
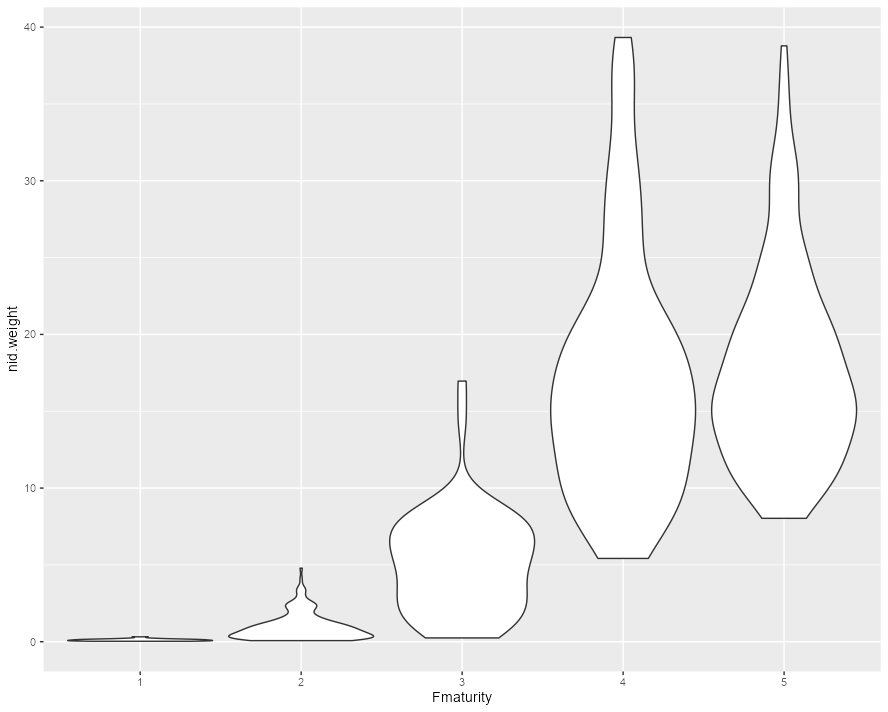
But my violin graphs are really thin, and they don't seem to help me visualize the data. So I tried using
ggplot(squid, aes(x=Fmaturity, y=nid.weight))
geom_violin(scale = "width")
which made the plots wider, except for the first violin plot. And what I'm wondering is:
when I use
scale = "width"is that changing the data that the graph is using. I don't want to manipulate the data, I just want to graph it so that I can see the violin plotsIs there a better way to show these violin plots so we can visualize the data at all stages? Can I make them wider without manipulating data?
CodePudding user response:
With scale = "width" the violins are all the same width:
ggplot(df, aes(x=Fmaturity, y=nid.weight))
geom_violin(scale = "width")
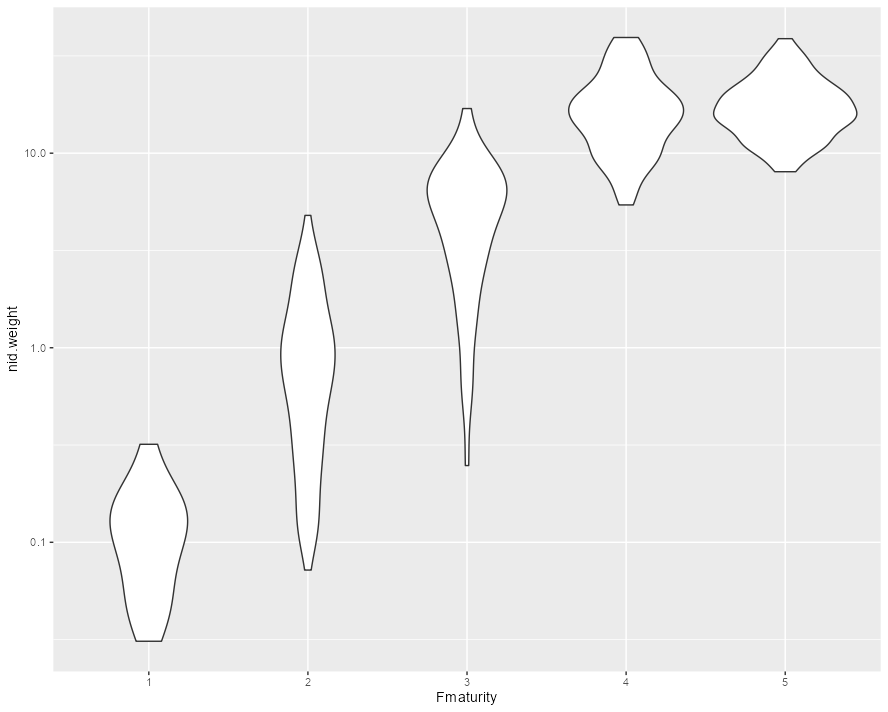
but the first violin is squashed vertically because...that's just the range of the data. If you want to be able to see the details of each distribution then you need a log scale on the y axis:
ggplot(df, aes(x=Fmaturity, y=nid.weight))
geom_violin()
scale_y_log10()