I will like to export my figure as a png or pdf or jpeg file in R. In simpler terms, I want to :
- include the export code as
png/pdf/jpegin the user-defined functiongraphplotter(please refer to the code below).
# dataset
Gen <- c('M','M','M','M','F','F','F','F','M','M','M','M','F','F','F','F')
Site <- rep('FRX',length(Gen))
Type <- c('L','L','L','L','L','L','L','L','R','R','R','R','R','R','R','R')
UID <- c(1001,1002,1003,1004,1001,1002,1003,1004,1001,1002,1003,1004,1001,1002,1003,1004)
Time_1 <- c(100.78,112.34,108.52,139.19,149.02,177.77,79.18,89.10,106.78,102.34,128.52,119.19,129.02,147.77,169.18,170.11)
Time_2 <- c(150.78,162.34,188.53,197.69,208.07,217.76,229.48,139.51,146.87,182.54,189.57,199.97,229.28,247.73,269.91,249.19)
Time_3 <- c(250.78,262.34,288.53,297.69,308.07,317.7,329.81,339.15,346.87,382.54,369.59,399.97,329.28,347.73,369.91,349.19)
Time_4 <- c(240.18,232.14,258.53,276.69,338.07,307.74,359.16,339.25,365.87,392.48,399.97,410.75,429.08,448.39,465.15,469.33)
Time_5 <-c(270.84,282.14,298.53,306.69,318.73,327.47,369.63,389.59,398.75,432.18,449.78,473.55,494.85,509.39,515.52,539.23)
df <- data.frame(Gen,Site,Type,UID,Time_1,Time_2,Time_3,Time_4,Time_5)
df
######################################################################################
# I know exporting as png looks something like this:
png(filename="turbidity-vs-time.png")
dev.off()
#####################################################################################
# I will like to include the 'exporting as png file code' in the "graphplotter" function below
##################################################################################
# user defined code to plot multiple graphs
# the graphplotter function below generates a graph, all I want is to export the output as a png file
library(tidyr)
library(ggplot2)
graphplotter <-function(x){
x %>%
gather(., time, turbidity, Time_1:Time_5, factor_key=TRUE) %>%
mutate(label = (paste0(Gen, "-", Type))) %>%
#group_by(UID) %>%
ggplot(aes(color = label)) geom_point(aes(time, turbidity, shape = label, group = label))
geom_line(aes(time, turbidity, group = label)) facet_wrap(~UID) theme(
legend.position = c(1, 1),
legend.justification = c("right", "top"),
legend.box.just = "right",
legend.margin = margin(1, 1, 1, 1),
legend.text = element_text(size = 7))
#1. export as png file code
}
graphplotter(df)
Update:
@David
 Thanks in advance!
Thanks in advance!
CodePudding user response:
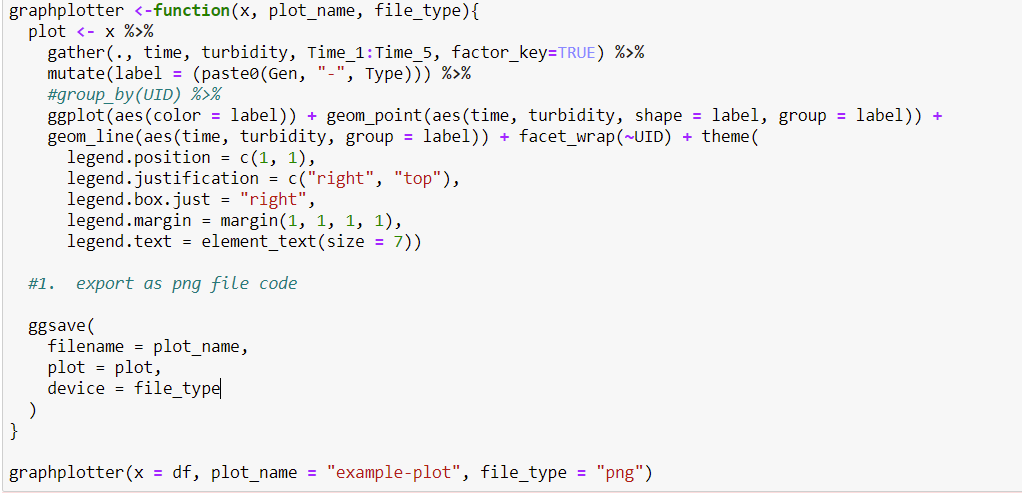
You could use ggplot2::ggsave() like so
graphplotter <-function(x, plot_name, file_type){
plot <- x %>%
gather(., time, turbidity, Time_1:Time_5, factor_key=TRUE) %>%
mutate(label = (paste0(Gen, "-", Type))) %>%
#group_by(UID) %>%
ggplot(aes(color = label)) geom_point(aes(time, turbidity, shape = label, group = label))
geom_line(aes(time, turbidity, group = label)) facet_wrap(~UID) theme(
legend.position = c(1, 1),
legend.justification = c("right", "top"),
legend.box.just = "right",
legend.margin = margin(1, 1, 1, 1),
legend.text = element_text(size = 7))
#1. export as png file code
ggsave(
filename = plot_name,
plot = plot,
device = file_type
)
}
The device argument will let you specify file type (be it .png, or .jpeg, etc.)
