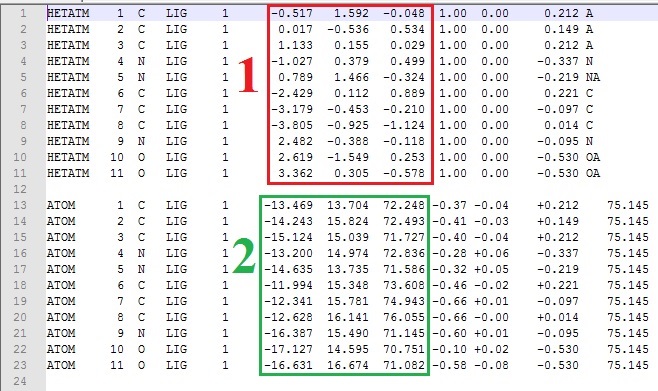
how's it going everybody ? I have a text file that contains data arranged in columns, and i need to replace some columns with others, and to be specific, xyz coordinates. what i'm looking for is described in the image below.(replace the red rectangle number 1 with the green rectangle number 2).
Many thanks in advance
HETATM 1 C LIG 1 -0.517 1.592 -0.048 1.00 0.00 0.212 A
HETATM 2 C LIG 1 0.017 -0.536 0.534 1.00 0.00 0.149 A
HETATM 3 C LIG 1 1.133 0.155 0.029 1.00 0.00 0.212 A
HETATM 4 N LIG 1 -1.027 0.379 0.499 1.00 0.00 -0.337 N
HETATM 5 N LIG 1 0.789 1.466 -0.324 1.00 0.00 -0.219 NA
HETATM 6 C LIG 1 -2.429 0.112 0.889 1.00 0.00 0.221 C
HETATM 7 C LIG 1 -3.179 -0.453 -0.210 1.00 0.00 -0.097 C
HETATM 8 C LIG 1 -3.805 -0.925 -1.124 1.00 0.00 0.014 C
HETATM 9 N LIG 1 2.482 -0.388 -0.118 1.00 0.00 -0.095 N
HETATM 10 O LIG 1 2.619 -1.549 0.253 1.00 0.00 -0.530 OA
HETATM 11 O LIG 1 3.362 0.305 -0.578 1.00 0.00 -0.530 OA
ATOM 1 C LIG 1 -13.469 13.704 72.248 -0.37 -0.04 0.212 75.145
ATOM 2 C LIG 1 -14.243 15.824 72.493 -0.41 -0.03 0.149 75.145
ATOM 3 C LIG 1 -15.124 15.039 71.727 -0.40 -0.04 0.212 75.145
ATOM 4 N LIG 1 -13.200 14.974 72.836 -0.28 0.06 -0.337 75.145
ATOM 5 N LIG 1 -14.635 13.735 71.586 -0.32 0.05 -0.219 75.145
ATOM 6 C LIG 1 -11.994 15.348 73.608 -0.46 -0.02 0.221 75.145
ATOM 7 C LIG 1 -12.341 15.781 74.943 -0.66 0.01 -0.097 75.145
ATOM 8 C LIG 1 -12.628 16.141 76.055 -0.66 -0.00 0.014 75.145
ATOM 9 N LIG 1 -16.387 15.490 71.145 -0.60 0.01 -0.095 75.145
ATOM 10 O LIG 1 -17.127 14.595 70.751 -0.10 0.02 -0.530 75.145
ATOM 11 O LIG 1 -16.631 16.674 71.082 -0.58 -0.08 -0.530 75.145
CodePudding user response:
It's not clear if you are trying to align the rows contextually, but if you are literally just wanting to replace columns 6, 7, and 8 with the columns from the same row in the other file, you can just do something like:
$ cat file1
HETATM 1 C LIG 1 -0.517 1.592 -0.048 1.00 0.00 0.212 A
HETATM 2 C LIG 1 0.017 -0.536 0.534 1.00 0.00 0.149 A
HETATM 3 C LIG 1 1.133 0.155 0.029 1.00 0.00 0.212 A
HETATM 4 N LIG 1 -1.027 0.379 0.499 1.00 0.00 -0.337 N
HETATM 5 N LIG 1 0.789 1.466 -0.324 1.00 0.00 -0.219 NA
HETATM 6 C LIG 1 -2.429 0.112 0.889 1.00 0.00 0.221 C
HETATM 7 C LIG 1 -3.179 -0.453 -0.210 1.00 0.00 -0.097 C
HETATM 8 C LIG 1 -3.805 -0.925 -1.124 1.00 0.00 0.014 C
HETATM 9 N LIG 1 2.482 -0.388 -0.118 1.00 0.00 -0.095 N
HETATM 10 O LIG 1 2.619 -1.549 0.253 1.00 0.00 -0.530 OA
HETATM 11 O LIG 1 3.362 0.305 -0.578 1.00 0.00 -0.530 OA
$ cat file2
ATOM 1 C LIG 1 -13.469 13.704 72.248 -0.37 -0.04 0.212 75.145
ATOM 2 C LIG 1 -14.243 15.824 72.493 -0.41 -0.03 0.149 75.145
ATOM 3 C LIG 1 -15.124 15.039 71.727 -0.40 -0.04 0.212 75.145
ATOM 4 N LIG 1 -13.200 14.974 72.836 -0.28 0.06 -0.337 75.145
ATOM 5 N LIG 1 -14.635 13.735 71.586 -0.32 0.05 -0.219 75.145
ATOM 6 C LIG 1 -11.994 15.348 73.608 -0.46 -0.02 0.221 75.145
ATOM 7 C LIG 1 -12.341 15.781 74.943 -0.66 0.01 -0.097 75.145
ATOM 8 C LIG 1 -12.628 16.141 76.055 -0.66 -0.00 0.014 75.145
ATOM 9 N LIG 1 -16.387 15.490 71.145 -0.60 0.01 -0.095 75.145
ATOM 10 O LIG 1 -17.127 14.595 70.751 -0.10 0.02 -0.530 75.145
ATOM 11 O LIG 1 -16.631 16.674 71.082 -0.58 -0.08 -0.530 75.145
$ awk '{getline s < "file2"; split(s, a); $6 = a[6]; $7 = a[7]; $8 = a[8]}1' file1
HETATM 1 C LIG 1 -13.469 13.704 72.248 1.00 0.00 0.212 A
HETATM 2 C LIG 1 -14.243 15.824 72.493 1.00 0.00 0.149 A
HETATM 3 C LIG 1 -15.124 15.039 71.727 1.00 0.00 0.212 A
HETATM 4 N LIG 1 -13.200 14.974 72.836 1.00 0.00 -0.337 N
HETATM 5 N LIG 1 -14.635 13.735 71.586 1.00 0.00 -0.219 NA
HETATM 6 C LIG 1 -11.994 15.348 73.608 1.00 0.00 0.221 C
HETATM 7 C LIG 1 -12.341 15.781 74.943 1.00 0.00 -0.097 C
HETATM 8 C LIG 1 -12.628 16.141 76.055 1.00 0.00 0.014 C
HETATM 9 N LIG 1 -16.387 15.490 71.145 1.00 0.00 -0.095 N
HETATM 10 O LIG 1 -17.127 14.595 70.751 1.00 0.00 -0.530 OA
HETATM 11 O LIG 1 -16.631 16.674 71.082 1.00 0.00 -0.530 OA
CodePudding user response:
Assuming that files have the same length, you could merge them with paste. and then extract columns in the desired order:
paste file1.txt file2.txt|awk '{print $1, $2, $3, $4, $5, $18, $19, $20, $9, $10, $11, $12}'