I am a new baby in R, I would like to ask for help to make the boxplot with the group I have 2 files, file 1 is the value of the samples (gene expression) test.txt
| gene | group1.1 | group1.2 | group2.1 | group2.2 |
|---|---|---|---|---|
| a1 | 12 | 13 | 12 | 12 |
| a2 | 2 | 3 | 25 | 31 |
| a3 | 24 | 30 | 34 | 22 |
| a4 | 10 | 11 | 23 | 24 |
and file 2 is the sample design design.txt
| file | condition |
|---|---|
| group1.1 | group1 |
| group1.2 | group1 |
| group2.1 | group2 |
| group2.2 | group2 |
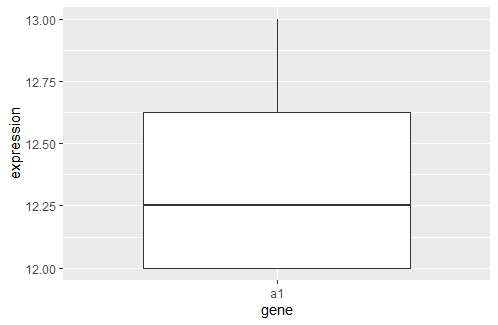
I want to make the boxplot in R with one specific row for example: a1 and have 2 groups 1, and 2; the output looks like boxplot-a1
How can I do this, direct from 2 files? I think I do the stupid way
dt1 <- read.delim("test.txt", sep="\t", header = TRUE)
dg <- read.delim("design.txt", sep="\t", header = TRUE)
I make the new file by copy and transpose:
| gene | name | group | expression |
|---|---|---|---|
| a1 | Group1.1 | group1 | 12 |
| a1 | Group1.2 | group1 | 13 |
| a1 | Group2.1 | group2 | 12 |
| a1 | Group2.2 | group2 | 12.5 |
| a2 | Group1.1 | group1 | 2 |
| a2 | Group1.2 | group1 | 3 |
| a2 | Group2.1 | group2 | 25 |
| a2 | Group2.2 | group2 | 31 |
dt <- read.delim("test_t.csv", sep="\t", header = TRUE)
a1 <- dt[dt$gene %in% "a1",]
ggplot(a1, aes(x=a1$group, y=a1$expression))
labs(title = "Expression A1", x = "Group", y = "Expression")
stat_boxplot(geom = "errorbar", width = 0.15)
geom_boxplot()
Thank you so much for your help!
CodePudding user response:
Having such data, it is worth first converting the variables of the chr type to factor.
library(tidyverse)
df = read.table(
header = TRUE,text="
gene name group expression
a1 Group1.1 group1 12
a1 Group1.2 group1 13
a1 Group2.1 group2 12
a1 Group2.2 group2 12.5
a2 Group1.1 group1 2
a2 Group1.2 group1 3
a2 Group2.1 group2 25
a2 Group2.2 group2 31") %>%
as_tibble() %>%
mutate(
gene = gene %>% fct_inorder(),
name = name %>% fct_inorder(),
group = group %>% fct_inorder()
)
Now you can make a boxplot for one value of the gene variable
df %>% filter(gene == "a1") %>%
ggplot(aes(gene, expression))
geom_boxplot()
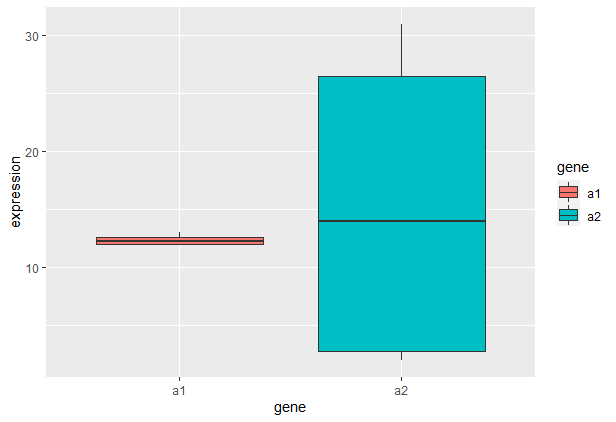
Either for both values at once
df %>%
ggplot(aes(gene, expression, fill=gene))
geom_boxplot()