I would like to add 2 different regression curves, coming from different models, in a scatter plot. Let's use the example below:
Weight=c(12.6,12.6,16.01,17.3,17.7,10.7,17,10.9,15,14,13.8,14.5,17.3,10.3,12.8,14.5,13.5,14.5,17,14.3,14.8,17.5,2.9,21.4,15.8,40.2,27.3,18.3,10.7,0.7,42.5,1.55,46.7,45.3,15.4,25.6,18.6,11.7,28,35,17,21,41,42,18,33,35,19,30,42,23,44,22)
Increment=c(0.55,0.53,16.53,55.47,80,0.08,41,0.1,6.7,2.2,1.73,3.53,64,0.05,0.71,3.88,1.37,3.8,40,3,26.3,29.7,10.7,35,27.5,60,43,31,21,7.85,63,9.01,67.8,65.8,27,40.1,31.2,22.3,35,21,74,75,12,19,4,20,65,46,9,68,74,57,57)
Id=c(rep("Aa",20),rep("Ga",18),rep("Za",15))
df=data.frame(Id,Weight,Increment)
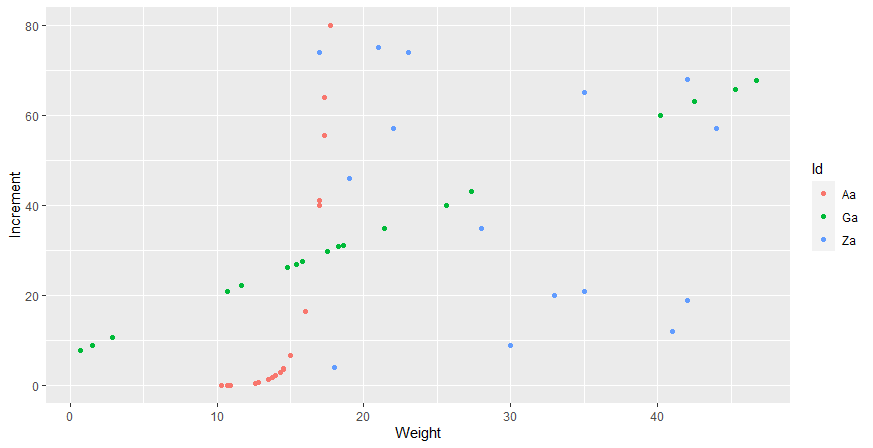
The scatter plot looks like this:
plot_df <- ggplot(df, aes(x = Weight, y = Increment, color=Id)) geom_point()
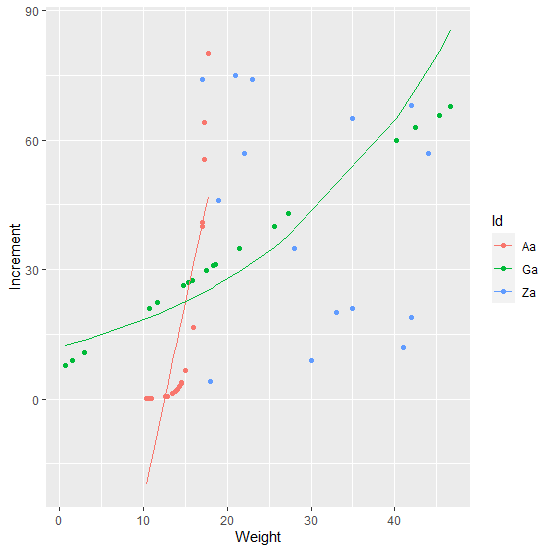
I tested a linear and an exponential regression model and could extract the results following loki's answer 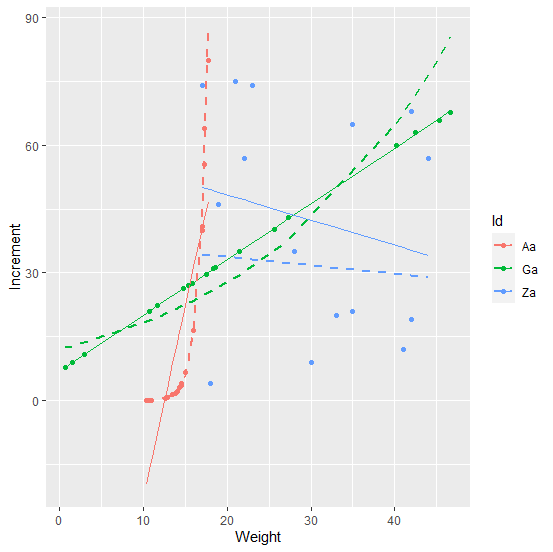
To only show the Aa fitted lines for the linear model and Ga fitted lines for the exponential model NA out the portions not wanted. In this case we used solid lines for the fitted models.
df %>%
ggplot(aes(Weight, Increment, color=Id))
geom_point()
geom_line(aes(y = ifelse(Id == "Aa", fitted(fm.lin), NA)))
geom_line(aes(y = ifelse(Id == "Ga", exp(fitted(fm.exp)), NA)))
CodePudding user response:
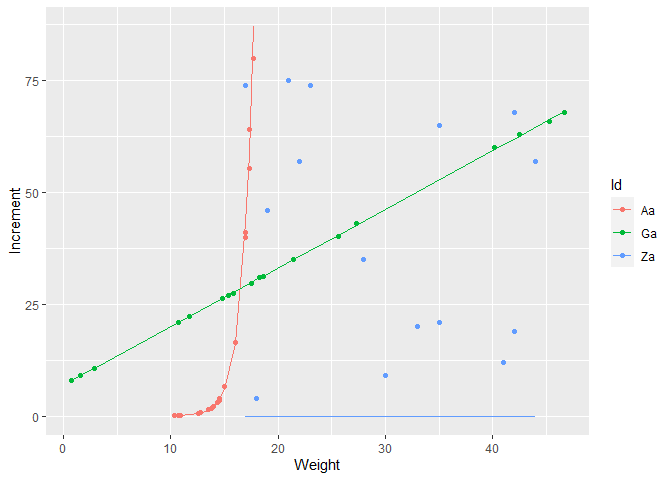
possible solution
Weight=c(12.6,12.6,16.01,17.3,17.7,10.7,17,10.9,15,14,13.8,14.5,17.3,10.3,12.8,14.5,13.5,14.5,17,14.3,14.8,17.5,2.9,21.4,15.8,40.2,27.3,18.3,10.7,0.7,42.5,1.55,46.7,45.3,15.4,25.6,18.6,11.7,28,35,17,21,41,42,18,33,35,19,30,42,23,44,22)
Increment=c(0.55,0.53,16.53,55.47,80,0.08,41,0.1,6.7,2.2,1.73,3.53,64,0.05,0.71,3.88,1.37,3.8,40,3,26.3,29.7,10.7,35,27.5,60,43,31,21,7.85,63,9.01,67.8,65.8,27,40.1,31.2,22.3,35,21,74,75,12,19,4,20,65,46,9,68,74,57,57)
Id=c(rep("Aa",20),rep("Ga",18),rep("Za",15))
df=data.frame(Id,Weight,Increment)
library(tidyverse)
df_model <- df %>%
group_nest(Id) %>%
mutate(
formula = c(
"lm(log(Increment) ~ Weight, data = .x)",
"lm(Increment ~ Weight,data = .x)",
"lm(Increment ~ 0,data = .x)"
),
transform = c("exp(fitted(.x))",
"fitted(.x)",
"fitted(.x)")
) %>%
mutate(model = map2(data, formula, .f = ~ eval(parse(text = .y)))) %>%
mutate(fit = map2(model, transform, ~ eval(parse(text = .y)))) %>%
select(Id, data, fit) %>%
unnest(c(data, fit))
ggplot(df_model)
geom_point(aes(Weight, Increment, color = Id))
geom_line(aes(Weight, fit, color = Id))
Created on 2021-10-06 by the reprex package (v2.0.1)