I'm using Regex in R and I am trying to capture a specific measurement of cardiac wall thickness that has 1-2 digits and 0-2 decimals as in:
"maximum thickness of lv wall= 1.5"
yet I want to exclude instances where (after|myectomy|resection) is somewhere after the word "thickness"
So I wrote the following regex code:
pattern <- "(?i)(?<=thickness)(?!(\\s{0,10}[[:alpha:]]{1,100}){0,8}\\s{0,10}(after|myectomy|resection))(?:(?:\\s{0,10}[[:alpha:]]{0,100}){0,8}\\s{0,10}[:=\\(]?)\\d{1,3}\\.?\\d{0,3}"
you can test it against this sample dataframe (every measurement in this example should match, except the last one):
df <- tibble(
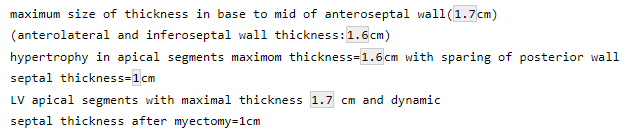
test = c("maximum size of thickness in base to mid of anteroseptal wall(1.7cm)",
"(anterolateral and inferoseptal wall thickness:1.6cm)",
"hypertrophy in apical segments maximom thickness=1.6cm with sparing of posterior wall",
"septal thickness=1cm",
"LV apical segments with maximal thickness 1.7 cm and dynamic",
"septal thickness after myectomy=1cm")
)
this regex code works for Matching what I want; the problem is that here I want to capture the measurements only, yet the sections behind the measurement are also getting captured although I have stated otherwise through none-capturing groups ?: .
Check this image out that is a result of stringr::str_view(df$test, pattern):
CodePudding user response:
You can use
pattern <- "(?i)(?<=\\bthickness(?:\\s{1,10}(?!(?:after|myectomy|resection)\\b)[a-zA-Z]{1,100}){0,8}\\s{0,10}[:=(]?)\\d{1,3}(?:\\.\\d{1,3})?"
str_view(df$test, pattern)
Output:
See the regex demo (JavaScript engine in modern browsers supports unlimited length lookbehind).
Details:
(?<=- start of the positive lookbehind that requires the following sequence of patterns to match immediately to the left of the current location:\bthickness- whole wordthickness(?:\s{1,10}(?!(?:after|myectomy|resection)\b)[a-zA-Z]{1,100}){0,8}- zero to eight occurrences of\s{1,10}- one to ten whitespaces(?!(?:after|myectomy|resection)\b)- noafter,mectomyandresectionwords are allowed immediately to the right of the current location[a-zA-Z]{1,100}- 1 to 100 ASCII letters
\s{0,10}- zero to ten whitespaces[:=(]?- an optional:,=or(char
)- end of the positive lookbehind\d{1,3}- one to three digits(?:\.\d{1,3})?- an optional sequence of a.and then one to three digits.
CodePudding user response:
Maybe you break it down to smaller steps.
tt <- regmatches(s, regexpr("(?i)thickness.*?\\d{1,3}(\\.\\d{1,3})?", s, perl = TRUE))
is.na(tt) <- grep("after|myectomy|resection", tt)
sub("[^0-9]*", "", tt)
#[1] "1.7" "1.6" "1.6" "1" "1.7" NA
Data:
s <- c("maximum size of thickness in base to mid of anteroseptal wall(1.7cm)",
"(anterolateral and inferoseptal wall thickness:1.6cm)",
"hypertrophy in apical segments maximom thickness=1.6cm with sparing of posterior wall",
"septal thickness=1cm",
"LV apical segments with maximal thickness 1.7 cm and dynamic",
"septal thickness after myectomy=1cm")