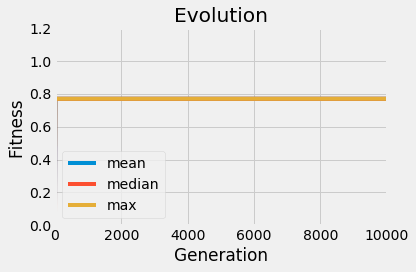
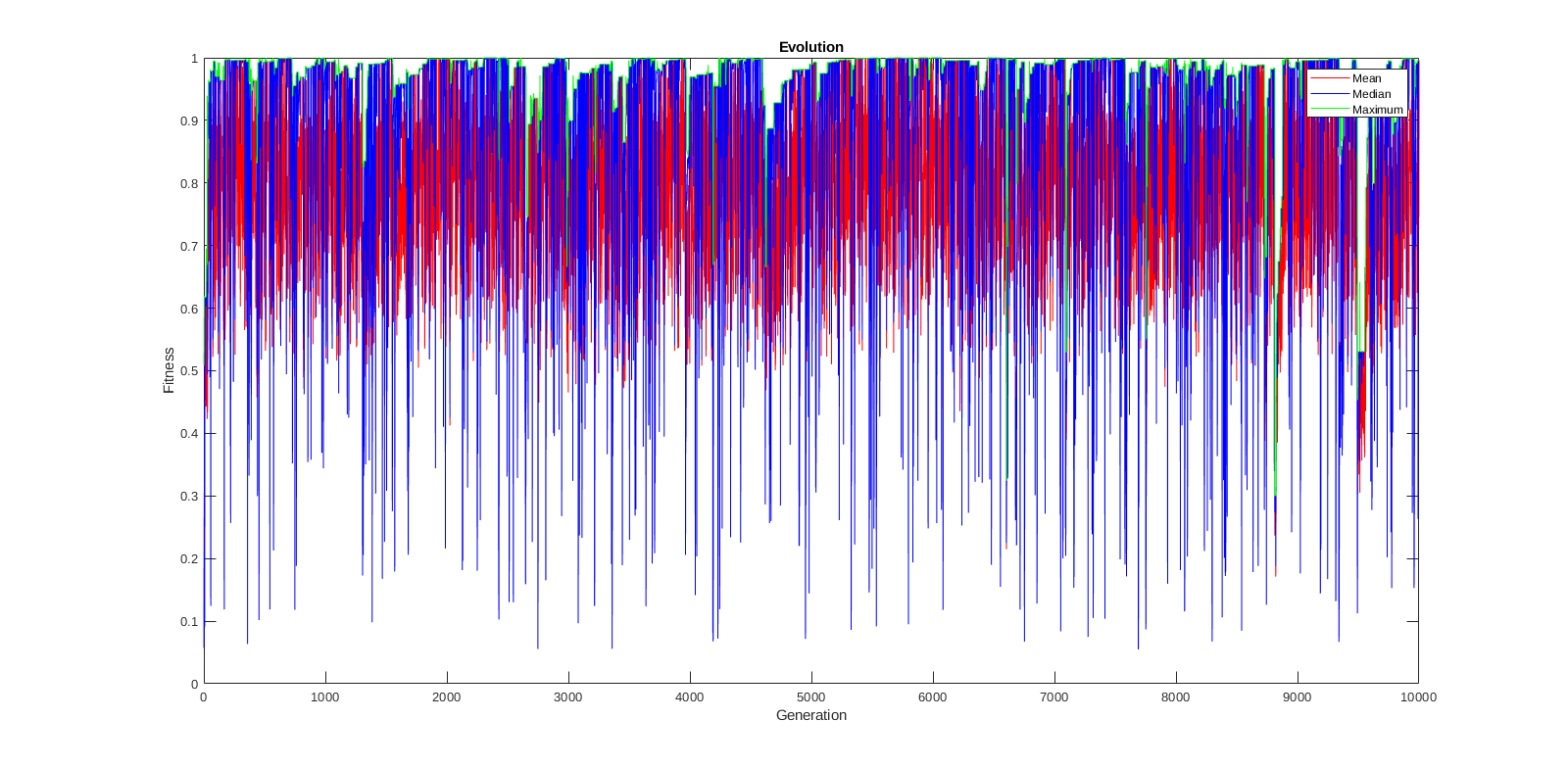
I want to port the working matlab code of a GA to python. In matlab I get to optimum within a 10% margin (good enough for a quick glance) with a population of 10 and 10K generations. Now I tried to port this code to python and get the odd behaviour that the solution seems stuck on a specific (but random) value sometimes way too far from the optimum.
A call of example1p6A(10000,10,0,0) using the provided matlab code results in
x*=
1.9707
3.0169
f(x*)=
0.9959
with some variance since it uses random numbers but is always close to above result. When you've an open figure it will draw the evolution over the gererations with the mean wiggling around 0.8.
HOWEVER when I call the same function in my ported python code I get variing results like
x* = [1.89979667 3.39332522]
f_max = 0.5499656257996797
all over the place sometimes way off. What makes me wonder is that the plotted min, max, and median seem to be all the same after a few generations in stark contrast to the matlab plots.
I've spent the whole day trying to find my error but had no success.
Stuff I've been aware of are:
- The indexes of vectors are starting from 0->N-1 in python instead of 1->N in matlab
- The one for loop is explicitly floored since I got an error for uneven populations. Matlab seems to do this implicitly.
- The equivalent to matlab
size()in python isshape()
Now I'm out of ideas what might be the issue. I'll provide two plots so you can get a quick look at my results.
function example1p6A(NG,Np,rf,pf)
% example1p6A is a function to generate example 1.6A in
% Power Magnetic Devices: A Multi-Ojective Design Approach
% by S.D. Sudhoff
%
% Call:
% example1p6A(NG,Np,rf,pf)
%
% Input:
% NG = number of generations
% Np = size of population
% rf = report flag (set to 1 to write report)
% pf = plot flag (set to 1 to plot results)
%
% Output:
% All output is textual or graphical
%
% Internal:
% GA = structure of genetic algorithm parameters
% GA.Ng = number of genes
% GA.Np = number in population
% GA.NG = number of generations
% GA.pc = probability of crossover
% GA.pm = probabiligy of gene mutation
% GA.xmn = vector of minimum values for unnormalized genes
% GA.xmx = vector of maximum values for unnormalized genes
% g = generation counter
% P = the population (number of genes by number of individuals)
% PC = copy of the population
% x = uncoded gene values of population
% f = fitness of population
% M = mutated populatin
% fmn = mean fitness of population
% fmx = maximum fitness of population
% fmd = median fitness of population
%
% Version/Date:
% May 7, 2013
%
% Written by:
% S.D. Sudhoff
% Purdue University
% Electrical Engineering Building
% 465 Northwestern Avenue
% West Lafayette, IN 47907-2035
% [email protected]
% 765-497-7648
GA.Ng=2; % number of genes
GA.Np=Np; % size of population
GA.NG=NG; % number of generations
GA.pc=0.5; % probability of crossover
GA.alpha=0.5; % blend ratio for crossover
GA.pm=0.1; % probability of a gene being mutated
GA.xmn=[0 0]; % vector of minimum values for unnormalized genes
GA.xmx=[5 5]; % vector of maximum values for unnormalized genes
% initialize the population
P=rand(GA.Ng,GA.Np);
% loop over the generation
for g=1:GA.NG
% make copy of current population
PC=P;
% find the parameter values for each member of the population
x=decode(P,GA);
% find the fitness
f=fitness(x);
if (g<GA.NG)
% find the mating pool
M=select(P,f,GA);
% create the next generation
P=children(M,GA);
end
fmn(g)=mean(f);
fmx(g)=max(f);
fmd(g)=median(f);
% plot
if pf
figure(1)
s=mod(g-1,9) 1;
symbol={'o','x',' ','*','s','d','v','p','h'};
plot(x(1,:),x(2,:),symbol{s},'Color',[0.8 0.8 0.8]*(1-g/GA.NG));
hold on;
end
% text report
if rf
disp(['Generation = ' num2str(g)]);
disp('P=');
disp(PC)
disp('x=');
disp(x)
disp('f=');
disp(f)
if (g<GA.NG)
disp('M=');
disp(M);
end
disp(['mean f = ' num2str(fmn(g))]);
disp(['max f = ' num2str(fmx(g))]);
disp(['median f = ' num2str(fmd(g))]);
end
end % generation loop
% finish plots
% if (pf)
hold off
title('Parameter Values');
xlabel('x_1');
ylabel('x_2');
figure(2)
gen=1:GA.NG;
plot(gen,fmn,'r-',gen,fmd,'b',gen,fmx,'g');
legend('Mean','Median','Maximum');
xlabel('Generation');
ylabel('Fitness');
title('Evolution');
% end
% best fitness
[~,i]=max(f);
disp('x*=');
disp(x(:,i))
disp('f(x*)=');
disp(f(i));
end % example1p6A
function x=decode(P,GA)
% decode decodes genes to parameter values
%
% Inputs:
% P = population array
% GA = genetic algorithm parameters
%
% Outputs
% x = parameter array
x=zeros(size(P));
for g=1:GA.Ng
x(g,:)=GA.xmn(g) (GA.xmx(g)-GA.xmn(g))*P(g,:);
end
end
function f=fitness(x)
% fitness computes the fitness
%
% Inputs:
% x = parameter array
%
% Outputs
% f = fitness
x1=x(1,:);
x2=x(2,:);
f=1./((x1.*x2-6).^2 4*(x2-3).^2 1);
end
function M=select(P,f,GA)
% select determines the mating pool
%
% Inputs:
% P = population array
% f = fitness
% GA = genetic algorithm parameters
%
% Outputs:
% M = mating pool array
M=zeros(size(P));
l1=randi([1 GA.Np],GA.Np,1);
l2=randi([1 GA.Np],GA.Np,1);
for i=1:GA.Np
i1=l1(i);
i2=l2(i);
if (f(i1)>=f(i2))
M(:,i)=P(:,i1);
else
M(:,i)=P(:,i2);
end
end
end
function C=children(M,GA)
% children forms children from the mating pool
%
% Inputs:
% M = mating pool array
% GA = genetic algorithm parameters
%
% Outputs:
% C = array of children
% perform simple blend crossover
C=zeros(size(M));
for i=1:GA.Np/2
i2=2*i;
i1=i2-1;
if GA.pc>rand
mn=0.5*(M(:,i1) M(:,i2));
df=(M(:,i2)-M(:,i1))*GA.alpha*rand;
C(:,i1)=mn df;
C(:,i2)=mn-df;
else
C(:,i1)=M(:,i1);
C(:,i2)=M(:,i2);
end
end
% mutation
R=rand(GA.Ng,GA.Np);
index=GA.pm>rand(GA.Ng,GA.Np);
C(index)=R(index);
% gene repair
index=C>1;
C(index)=1;
index=C<0;
C(index)=0;
end
import numpy as np
import math, statistics
import matplotlib.pyplot as plt
def example1p6A(NG, Np, rf, pf):
# example1p6A is a function to generate example 1.6A in
# Power Magnetic Devices: A Multi-Ojective Design Approach
# by S.D. Sudhoff
#
# Call:
# example1p6A(NG,Np,rf,pf)
#
# Input:
# NG = number of generations
# Np = size of population
# rf = report flag (set to 1 to write report)
# pf = plot flag (set to 1 to plot results)
#
# Output:
# All output is textual or graphical
#
# Internal:
# GA = structure of genetic algorithm parameters
# GA.Ng = number of genes
# GA.Np = number in population
# GA.NG = number of generations
# GA.pc = probability of crossover
# GA.pm = probabiligy of gene mutation
# GA.xmn = vector of minimum values for unnormalized genes
# GA.xmx = vector of maximum values for unnormalized genes
# g = generation counter
# P = the population (number of genes by number of individuals)
# PC = copy of the population
# x = uncoded gene values of population
# f = fitness of population
# M = mutated populatin
# fmn = mean fitness of population
# fmx = maximum fitness of population
# fmd = median fitness of population
#
# Version/Date:
# May 7, 2013
#
# Written by:
# S.D. Sudhoff
# Purdue University
# Electrical Engineering Building
# 465 Northwestern Avenue
# West Lafayette, IN 47907-2035
# [email protected]
# 765-497-7648
GA = dict(
Np=np.array(Np),
NG=np.array(NG),
Ng=np.array(2),
pc=np.array(0.5),
alpha=np.array(0.5),
pm=np.array(0.1),
xmn=np.array([0, 0]),
xmx=np.array([5, 5]))
# Init population
P = np.random.rand(GA["Ng"],GA["Np"])
# empty bins fol plotting
fmean = []
fmax = []
fmed = []
# loop over the generations
for g in range(GA["NG"]):
PC = P
x = decode(P,GA)
f=fitness(x)
# if g < GA["NG"]: # g ist immer kleiner als GA["NG"] weil range() von 0->(n-1) geht
# M = select(P,f,GA)
# P = children(M,GA)
M = select(P,f,GA)
P = children(M,GA)
fmean.append(statistics.mean(f))
fmax.append(max(f))
fmed.append(statistics.median(f))
print("x* = {}".format(x[:,1]))
print("f_max = {}\n".format(f.max()))
# plot that stuff
plt.style.use("fivethirtyeight")
plt.plot(np.arange(1,GA["NG"] 1),fmean, label = "mean")
plt.plot(np.arange(1,GA["NG"] 1),fmed, label = "median")
plt.plot(np.arange(1,GA["NG"] 1),fmax, label = "max")
plt.xlabel("Generation")
plt.ylabel("Fitness")
plt.title("Evolution")
plt.axis([0,GA["NG"],0,1.2])
plt.tight_layout()
plt.legend()
plt.show()
# return P
def decode(P,GA):
x = np.zeros(np.shape(P))
for g in range(GA["Ng"]):
x[g,:] = GA["xmn"][g] (GA["xmx"][g]-GA["xmn"][g])*P[g,:]
return x
def fitness(x):
# fitness computes the fitness
#
# Inputs:
# x = parameter array
#
# Outputs
# f = fitness
x1 = x[0,:]
x2 = x[1,:]
f=1/((x1*x2-6)**2 4*(x2-3)**2 1)
return f
def select(P,f,GA):
# select determines the mating pool
#
# Inputs:
# P = population array
# f = fitness
# GA = genetic algorithm parameters
#
# Outputs:
# M = mating pool array
M = np.zeros(np.shape(P))
l1 = np.random.randint(1,GA["Np"] 1,(GA["Np"],1)) #randint is [lower...upper)
l2 = np.random.randint(1,GA["Np"] 1,(GA["Np"],1))
for i in range(GA["Np"]):
i1 = l1[i][0]
i2 = l2[i][0]
if f[i1-1] >= f[i2-1]:
#bei matlab beginnt der index bei 1. python hingegen startet bei 0 und endet bei n-1
M[:,i] = P[:,i1-1]
else:
M[:,i] = P[:,i2-1]
return M
def children(M,GA):
# children forms children from the mating pool
#
# Inputs:
# M = mating pool array
# GA = genetic algorithm parameters
# # Outputs:
# C = array of children
C = np.zeros(np.shape(M))
# perform simple blend crossover
C = np.zeros(np.shape(M))
for i in range(1,math.floor(GA["Np"]/2) 1):
i2 = 2*i
i1 = i2-1
rnd = np.random.rand()
if GA["pc"] > rnd:
mn= 0.5*(M[:,i1-1] M[:,i2-1]) #the index starts from 0 but the counter from 1
df=(M[:,i2-1]-M[:,i1-1])*GA["alpha"]*np.random.rand()
C[:,i1-1]=mn df
C[:,i2-1]=mn-df
else:
C[:,i1-1]=M[:,i1-1]
C[:,i2-1]=M[:,i2-1]
# Mutation
R=np.random.rand(GA["Ng"],GA["Np"])
index = GA["pm"] > np.random.rand(GA["Ng"],GA["Np"])
C[index]=C[index]
# Gene repair
index= C>1
C[index] = 1
index =C <0
C[index] = 0
return C
example1p6A(10000,10,0,0)
CodePudding user response:
Although I've found one error C[index] = C[index] where it should have been C[index]=R[index] I've compltetely ditched my above code and wrote it from scratch with plenty print()-commands to see what each step is doing. Now I have working code as follows:
import numpy as np
import matplotlib.pyplot as plt
import math, statistics
def mygen(NG,Np,rf,pf):
GA = dict(
Ng = np.array(2),
Np = np.array(Np),
NG = np.array(NG),
pc = np.array(0.5),
alpha = np.array(0.5),
pm = np.array(0.1),
xmn = np.array([0,0]),
xmx = np.array([5,5])
)
# Init population
P = np.random.rand(GA['Ng'],GA['Np'])
# print("Iinitial population:\n{}".format(P))
# Empty bins for median and stuff
fmean = []
fmed = []
fmax = []
index_of_max = 0
for g in range(GA['NG']):
# print("Generation {}:".format(g))
x = decode(P,GA)
# print("\n\n")
f = fitness(x)
# print("fitness f() for each individual:\n{}".format(f))
index_of_max = np.unravel_index(np.argmax(f,axis=None),f.shape)
fmean.append(np.mean(f))
fmed.append(np.median(f))
fmax.append(f[index_of_max])
# print("Worst fitness for Individual #{}:{}".format(index_of_min,f[index_of_min]))
# print("Best fitness for Individual #{}:{}".format(index_of_max,f[index_of_max]))
# Find the mating pool
M = select(P,f,GA)
# Create new population
P = children(M,GA)
# Final output
# index = np.unravel_index(np.argmax(f))
i = index_of_max
print("x*:{}".format(x[:,i]))
print("f(x*)={}".format(f[i]))
# Final plots
plt.style.use("fivethirtyeight")
plt.plot(np.arange(1,GA["NG"] 1),fmean,label = "mean")
plt.plot(np.arange(1,GA["NG"] 1),fmax,label = "max")
plt.plot(np.arange(1,GA["NG"] 1),fmed,label = "med")
plt.xlabel("Generation")
plt.ylabel("Fitness")
plt.title("Evolution")
plt.axis([0,GA["NG"],0,1.2])
# plt.xticks(np.arange(1,GA["NG"] 1, 5))
plt.tight_layout()
plt.legend()
plt.show()
def decode(P,GA):
x=np.zeros(np.shape(P))
for g in range(GA['Ng']):
x[g,:] = GA['xmn'][g] (GA['xmx'][g]-GA['xmn'][g])*P[g,:]
# print("decode()\n x{}:\n{}".format(g,x))
return x
def fitness(x):
x1 = x[0,:]
x2 = x[1,:]
f = 1/((x1*x2-6)**2 4*(x2-3)**2 1)
return f
def select(P,f,GA):
M = np.zeros(np.shape(P))
l1 = np.random.randint(0,GA["Np"],(GA["Np"],1))
l2 = np.random.randint(0,GA["Np"],(GA["Np"],1))
for i in range(GA["Np"]):
i1 = l1[i,0]
i2 = l2[i,0]
if f[i1] >= f[i2]:
M[:,i] = P[:,i1]
# print(f[i1])
else:
M[:,i] = P[:,i2]
# print(f[i2])
# print(i1)
return M
def children(M,GA):
C = np.zeros(np.shape(M))
# Simple blend crossover
for i in range(math.floor(GA["Np"]/2)):
i2 = 2*(i 1)
i1 = i2-1
if GA["pc"] > np.random.rand():
mn = 0.5*(M[:,i1-1] M[:,i2-1])
df = (M[:,i2-1]-M[:,i1-1])*GA["alpha"]*np.random.rand()
C[:,i1-1] = mn df
C[:,i2-1] = mn-df
else:
C[:,i1-1] = M[:,i1-1]
C[:,i2-1] = M[:,i2-1]
# Mutation
R = np.random.rand(GA["Ng"],GA["Np"])
index = GA["pm"]>np.random.rand(GA["Ng"],GA["Np"])
C[index] = R[index]
# Gene repair
index =C>1
C[index]=1
index =C<0
C[index]=0
return C
mygen(15,50,1,0)