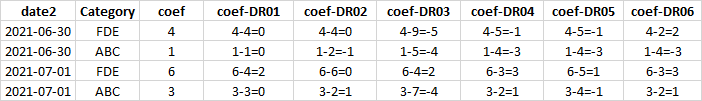
I would like to know how I can do a subtraction between the dataset I got (All) with my df1 dataset. I inserted an image to illustrate the output. So you'll notice that I want to subtract the value of the coef of All from the columns of DR0.. of the df1.
library(dplyr)
library(tidyverse)
library(lubridate)
df1 <- structure(
list(date1= c("2021-06-28","2021-06-28","2021-06-28","2021-06-28"),
date2 = c("2021-06-30","2021-06-30","2021-07-01","2021-07-01"),
Category = c("FDE","ABC","FDE","ABC"),
Week= c("Wednesday","Wednesday","Friday","Friday"),
DR1 = c(4,1,6,3),
DR01 = c(4,1,4,3), DR02= c(4,2,6,2),DR03= c(9,5,4,7),
DR04 = c(5,4,3,2),DR05 = c(5,4,5,4),
DR06 = c(2,4,3,2)),
class = "data.frame", row.names = c(NA, -4L))
> df1
date1 date2 Category Week DR1 DR01 DR02 DR03 DR04 DR05 DR06
1 2021-06-28 2021-06-30 FDE Wednesday 4 4 4 9 5 5 2
2 2021-06-28 2021-06-30 ABC Wednesday 1 1 2 5 4 4 4
3 2021-06-28 2021-07-01 FDE Friday 6 4 6 4 3 5 3
4 2021-06-28 2021-07-01 ABC Friday 3 3 2 7 2 4 2
return_coef <- function(dmda, CategoryChosse) {
x<-df1 %>% select(starts_with("DR0"))
x<-cbind(df1, setNames(df1$DR1 - x, paste0(names(x), "_PV")))
PV<-select(x, date2,Week, Category, DR1, ends_with("PV"))
med<-PV %>%
group_by(Category,Week) %>%
summarize(across(ends_with("PV"), median))
SPV<-df1%>%
inner_join(med, by = c('Category', 'Week')) %>%
mutate(across(matches("^DR0\\d $"), ~.x
get(paste0(cur_column(), '_PV')),
.names = '{col}_{col}_PV')) %>%
select(date1:Category, DR01_DR01_PV:last_col())
SPV<-data.frame(SPV)
mat1 <- df1 %>%
filter(date2 == dmda, Category == CategoryChosse) %>%
select(starts_with("DR0")) %>%
pivot_longer(cols = everything()) %>%
arrange(desc(row_number())) %>%
mutate(cs = cumsum(value)) %>%
filter(cs == 0) %>%
pull(name)
(dropnames <- paste0(mat1,"_",mat1, "_PV"))
SPV <- SPV %>%
filter(date2 == dmda, Category == CategoryChosse) %>%
select(-any_of(dropnames))
datas<-SPV %>%
filter(date2 == ymd(dmda)) %>%
group_by(Category) %>%
summarize(across(starts_with("DR0"), sum)) %>%
pivot_longer(cols= -Category, names_pattern = "DR0(. )", values_to = "val") %>%
mutate(name = readr::parse_number(name))
colnames(datas)[-1]<-c("Days","Numbers")
datas <- datas %>%
group_by(Category) %>%
slice((as.Date(dmda) - min(as.Date(df1$date1) [
df1$Category == first(Category)])):max(Days) 1) %>%
ungroup
mod <- nls(Numbers ~ b1*Days^2 b2,start = list(b1 = 0,b2 = 0),data = datas, algorithm = "port")
as.numeric(coef(mod)[2])
}
All<-cbind(df1 %>% select(date2, Category), coef = mapply(return_coef, df1$date2, df1$Category))
> All
date2 Category coef
1 2021-06-30 FDE 4
2 2021-06-30 ABC 1
3 2021-07-01 FDE 6
4 2021-07-01 ABC 3
Output I want
CodePudding user response:
This is a combination of join/merge (see How to join (merge) data frames (inner, outer, left, right), What's the difference between INNER JOIN, LEFT JOIN, RIGHT JOIN and FULL JOIN?) and an across mutate:
library(dplyr)
left_join(All, df1, by = c("date2", "Category")) %>%
mutate(across(starts_with("DR0"), ~ coef - .))
# date2 Category coef date1 Week DR1 DR01 DR02 DR03 DR04 DR05 DR06
# 1 2021-06-30 FDE 4 2021-06-28 Wednesday 4 0 0 -5 -1 -1 2
# 2 2021-06-30 ABC 1 2021-06-28 Wednesday 1 0 -1 -4 -3 -3 -3
# 3 2021-07-01 FDE 6 2021-06-28 Friday 6 2 0 2 3 1 3
# 4 2021-07-01 ABC 3 2021-06-28 Friday 3 0 1 -4 1 -1 1
Data
df1 <- structure(list(date1 = c("2021-06-28", "2021-06-28", "2021-06-28", "2021-06-28"), date2 = c("2021-06-30", "2021-06-30", "2021-07-01", "2021-07-01"), Category = c("FDE", "ABC", "FDE", "ABC"), Week = c("Wednesday", "Wednesday", "Friday", "Friday"), DR1 = c(4, 1, 6, 3), DR01 = c(4, 1, 4, 3), DR02 = c(4, 2, 6, 2), DR03 = c(9, 5, 4, 7), DR04 = c(5, 4, 3, 2), DR05 = c(5, 4, 5, 4), DR06 = c(2, 4, 3, 2)), class = "data.frame", row.names = c(NA, -4L))
All <- structure(list(date2 = c("2021-06-30", "2021-06-30", "2021-07-01", "2021-07-01"), Category = c("FDE", "ABC", "FDE", "ABC"), coef = c(4L, 1L, 6L, 3L)), class = "data.frame", row.names = c("1", "2", "3", "4"))
CodePudding user response:
Here is an alternative dplyr way: We use bind_cols a rare used dplyr function. Interestingly without transforming to integer class it did not work:
library(dplyr)
df1 %>%
bind_cols(coef = All$coef) %>%
mutate(across(DR1:coef, as.integer)
across(DR01:DR06, ~coef - .))
date1 date2 Category Week DR1 DR01 DR02 DR03 DR04 DR05 DR06 coef
<chr> <chr> <chr> <chr> <int> <int> <int> <int> <int> <int> <int> <int>
1 2021-06-28 2021-06-30 FDE Wednesday 4 0 0 -5 -1 -1 2 4
2 2021-06-28 2021-06-30 ABC Wednesday 1 0 -1 -4 -3 -3 -3 1
3 2021-06-28 2021-07-01 FDE Friday 6 2 0 2 3 1 3 6
4 2021-06-28 2021-07-01 ABC Friday 3 0 1 -4 1 -1 1 3