This is my code:
with open('file.txt','r') as f:
table = {}
for lines in f:
co1,co2,co3,pro1,pro2,pro3 = (
item.strip() for item in lines.split(',',5))
codon=str(co1 co2 co3)
table[codon] = pro2
print(table)
I need to split the dan_seq whatever they say it is into chunks of 3 and then match it to the dictionary I created
i.e. if the chunk is a key in my dictionary, return the value of it
dna_seq = list('AAAGTTAAATAATAAATAGGTGAA')
the picture is the text file:
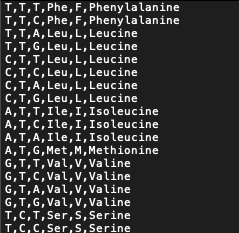
CodePudding user response:
You can use slicing to chunk up the dna_seq, and then table.get to get the matching values:
>>> dna_seq = 'AAAGTTAAATAATAAATAGGTGAA'
>>> [dna_seq[i:i 3] for i in range(0, len(dna_seq), 3)]
['AAA', 'GTT', 'AAA', 'TAA', 'TAA', 'ATA', 'GGT', 'GAA']
>>> for chunk in [dna_seq[i:i 3] for i in range(0, len(dna_seq), 3)]:
... print(table.get(chunk))
CodePudding user response:
You need to split the string into chunks of size 3, which you can accomplish by: dna_seq[i:i 3] for i in range(0, len(dna_seq), 3) and your final code would look somewhat like:
for i in range(0, len(dna_seq), 3):
current_seq = dna_seq[i:i 3]
# check if this value exists, then print it
if current_seq in table:
print(table[current_seq])
