I am currently working with 30 datasets with the same column names, but different numeric data. I need to apply a linear mixed model and a generalised linear model to every instance of the dataset and plot the resulting fixed effect coefficients on a forest plot.
The data is currently structured as follows (using the same dataset for every list element for simplicity):
library(lme4)
data_list <- list()
# There's definitely a better way of doing this through lapply(), I just can't figure out how
for (i in 1:30){
data_list[[i]] <- tibble::as_tibble(mtcars) # this would originally load different data at every instance
}
compute_model_lmm <- function(data){
lmer("mpg ~ hp disp drat (1|cyl)", data = data)
}
result_list_lmm <- lapply(data_list, compute_model_lmm)
What I am currently doing is
library(modelsummary)
modelplot(result_list_lmm)
facet_wrap(~model) #modelplot() takes arguments/functions from ggplot2
which takes an awful amount of time, but it works.
Now, I would like to compare another model on the same plot, as in
compute_model_glm <- function(data){
glm("mpg ~ hp disp drat cyl", data = data)
}
result_list_glm <- lapply(data_list, compute_model_glm)
modelplot(list(result_list_lmm[[1]], result_list_glm[[1]]))
but for every instance of the plot.
How do I specify it to modelplot()?
Thanks in advance!
CodePudding user response:
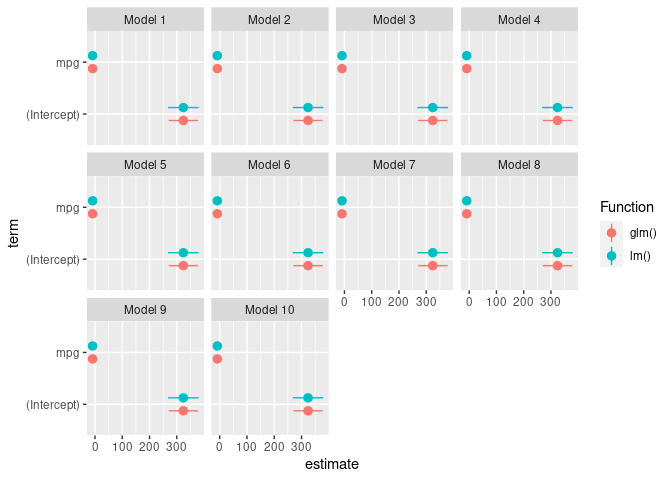
The modelplot function gives you a few basic ways of plotting coefficients and intervals (check the facet argument, for example).
However, the real power of the function comes by using the draw=FALSE argument. In that case, modelplot does the hard job of giving you the estimates in a convenient data frame, with all the renaming, robust standard errors, and other utilities of the modelplot function. Then, you can use that data frame to do the plotting yourself with ggplot2 for infinite customization.
library(modelsummary)
library(ggplot2)
results_lm <- lapply(1:10, function(x) lm(hp ~ mpg, data = mtcars)) |>
modelplot(draw = FALSE) |>
transform("Function" = "lm()")
results_glm <- lapply(1:10, function(x) glm(hp ~ mpg, data = mtcars)) |>
modelplot(draw = FALSE) |>
transform("Function" = "glm()")
results <- rbind(results_lm, results_glm)
head(results)
term model estimate std.error conf.low conf.high Function
1 (Intercept) Model 1 324.0823 27.4333 268.056 380.1086 lm()
3 (Intercept) Model 2 324.0823 27.4333 268.056 380.1086 lm()
5 (Intercept) Model 3 324.0823 27.4333 268.056 380.1086 lm()
7 (Intercept) Model 4 324.0823 27.4333 268.056 380.1086 lm()
9 (Intercept) Model 5 324.0823 27.4333 268.056 380.1086 lm()
11 (Intercept) Model 6 324.0823 27.4333 268.056 380.1086 lm()
ggplot(results, aes(y = term, x = estimate, xmin = conf.low, xmax = conf.high))
geom_pointrange(aes(color = Function), position = position_dodge(width = .5))
facet_wrap(~model)