I am trying to work on the 4 dimensional chaotic attractor Lyapunov spectrum and there values so far the code mention below works well for three dimensional system but errors arise in 4D and 5D system
import matplotlib.pyplot as plt
import numpy as np
from scipy.integrate import odeint
def diff_Lorenz(u):
x,y,z,w= u
f = [a*(y-x) , x*z w, b-x*y, z*y-c*w]
Df = [[-a,a,0,0], [z,0, x,1], [-y, -x, 0,0],[0,z,y,-c]]
return np.array(f), np.array(Df)
def LEC_system(u):
#x,y,z = u[:3]
U = u[2:18].reshape([4,4])
L = u[12:15]
f,Df = diff_Lorenz(u[:4])
A = U.T.dot(Df.dot(U))
dL = np.diag(A).copy();
for i in range(4):
A[i,i] = 0
for j in range(i 1,4): A[i,j] = -A[j,i]
dU = U.dot(A)
return np.concatenate([f,dU.flatten(),dL])
a=6;b=11;c=5;
u0 = np.ones(4)
U0 = np.identity(4)
L0 = np.zeros(4)
u0 = np.concatenate([u0, U0.flatten(), L0])
t = np.linspace(0,10,301)
u = odeint(lambda u,t:LEC_system(u),u0,t, hmax=0.05)
L = u[5:,12:15].T/t[5:]
# plt.plot(t[5:],L.T)
# plt.show()
p1=L[0,:];p2=L[1,:];p3=L[2,:];p4=L[3,:]
L1 = np.mean(L[0,:]);L2=np.average(L[1,:]);L3=np.average(L[2,:]);L4=np.average(L[3,:])
t1 = np.linspace(0,100,len(p1))
plt.plot(t1,p1);plt.plot(t1,p2);plt.plot(t1,p3);plt.plot(t1,p4)
# plt.show()
print('LES= ',L1,L2,L3,L4)
the output error is
D:\anaconda3\lib\site-packages\scipy\integrate\odepack.py:247: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.
warnings.warn(warning_msg, ODEintWarning)
---------------------------------------------------------------------------
IndexError Traceback (most recent call last)
~\AppData\Local\Temp/ipykernel_7008/1971199288.py in <module>
32 # plt.plot(t[5:],L.T)
33 # plt.show()
---> 34 p1=L[0,:];p2=L[1,:];p3=L[2,:];p4=L[3,:]
35 L1=np.mean(L[0,:]);L2=np.average(L[1,:]);L3=np.average(L[2,:]);L4=np.average(L[3,:])
36 t1 = np.linspace(0,100,len(p1))
IndexError: index 3 is out of bounds for axis 0 with size 3
what is wrong?
output expected is L1=.5162,L2=-.0001,L3=-4.9208,L4=-6.5954
CodePudding user response:
In LEC_system(u), the flat vector u contains in sequence
- the state vector,
ncomponents, - the eigenbasis
U, an x nmatrix - the accumulated exponents
L,ncomponents.
With n=4, this translates thus to the decomposition
def LEC_system(u):
#x,y,z,w = u[:4]
U = u[4:20].reshape([4,4])
L = u[20:24]
f,Df = diff_Lorenz(u[:4])
A = U.T.dot(Df.dot(U))
dL = np.diag(A).copy();
for i in range(4):
A[i,i] = 0
for j in range(i 1,4): A[i,j] = -A[j,i]
dU = U.dot(A)
return np.concatenate([f,dU.flatten(),dL])
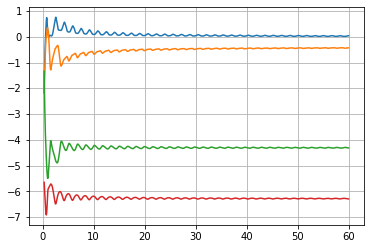
Of course, in the evaluation after the integration one has to likewise use the correct segment of the state vector
L = u[5:,20:24].T/t[5:]
and only using the latter quart of the graphs, after integrating to t=60
LES= 0.029214865425355396 -0.43816854013111833 -4.309199339754925 -6.28183676249535
This still are not the expected values, as that appears to be contracting along all directions transversal to the solution curve.