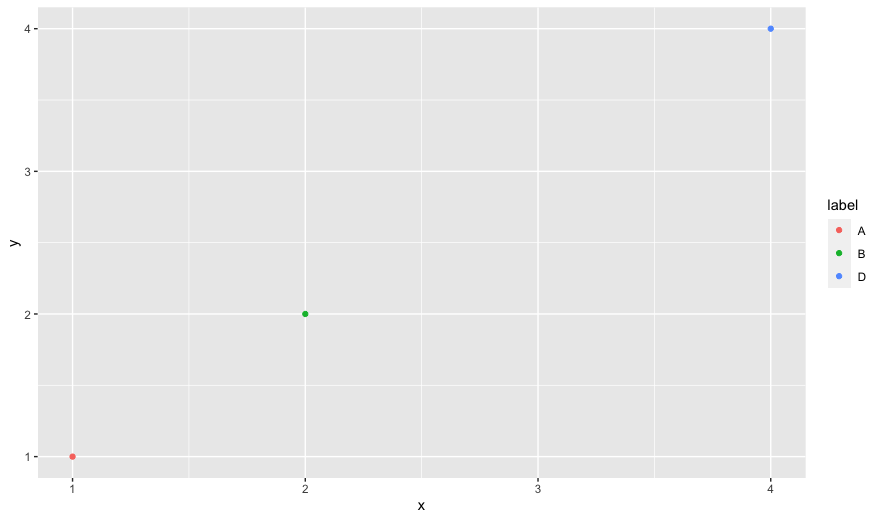
I think it is a recent change in ggplot2 that all the values defined in the scale_manual function are included in the legend, but I want only the values that are in the data set in the legend.
Below is a minimal example, in which "C" is filtered from the data set but still appears in the legend. Is there an easy fix to remove "C" from the legend?
library(ggplot2)
library(dplyr)
df_dummy <- tribble(
~label, ~x, ~y,
"A", 1, 1,
"B", 2, 2,
"C", 3, 3,
"D", 4, 4
)
myColors <- viridis::viridis_pal(option = "H")(4)
names(myColors) <- factor(df_dummy$label)
dummy_col_scale <- scale_color_manual(name = "Legend", values = myColors)
df_dummy %>%
filter(label != "C") %>%
ggplot(aes(x, y, color = label))
geom_point()
dummy_col_scale
sessionInfo()
R version 4.0.3 (2020-10-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19044)
Matrix products: default
locale:
[1] LC_COLLATE=English_Australia.1252
[2] LC_CTYPE=English_Australia.1252
[3] LC_MONETARY=English_Australia.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_Australia.1252
attached base packages:
[1] stats graphics grDevices utils
[5] datasets methods base
other attached packages:
[1] dplyr_1.0.5 ggplot2_3.3.5
loaded via a namespace (and not attached):
[1] rstudioapi_0.13 magrittr_2.0.1
[3] tidyselect_1.1.1 munsell_0.5.0
[5] cowplot_1.1.1 viridisLite_0.4.0
[7] colorspace_2.0-2 R6_2.5.1
[9] rlang_0.4.10 fansi_0.4.2
[11] tools_4.0.3 grid_4.0.3
[13] gtable_0.3.0 utf8_1.2.1
[15] cli_3.0.1 DBI_1.1.1
[17] withr_2.4.2 ellipsis_0.3.2
[19] digest_0.6.27 assertthat_0.2.1
[21] tibble_3.1.1 lifecycle_1.0.1
[23] crayon_1.4.1 gridExtra_2.3
[25] farver_2.1.0 purrr_0.3.4
[27] viridis_0.6.0 vctrs_0.3.8
[29] glue_1.4.2 labeling_0.4.2
[31] compiler_4.0.3 pillar_1.6.4
[33] generics_0.1.0 scales_1.1.1
[35] pkgconfig_2.0.3
CodePudding user response:
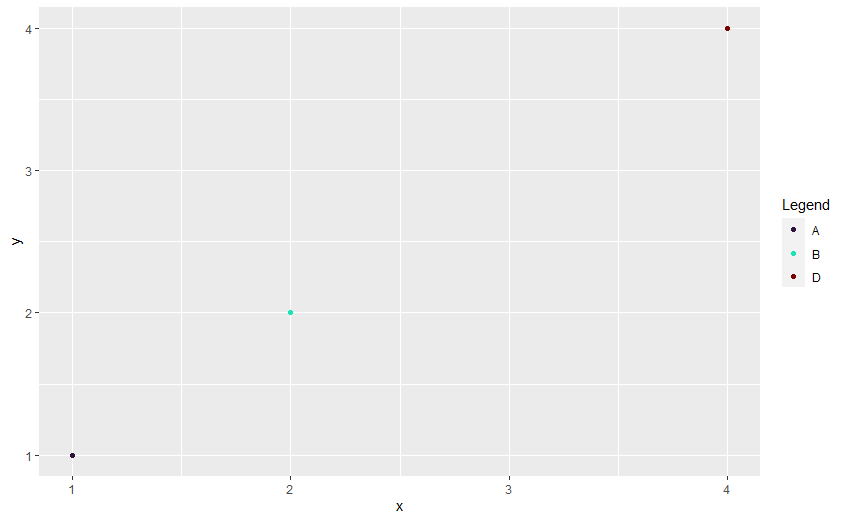
I believe there are a few solutions to your problem. One potential solution is to do the filtering first then select 3 colours:
library(ggplot2)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
df_dummy <- tribble(
~label, ~x, ~y,
"A", 1, 1,
"B", 2, 2,
"C", 3, 3,
"D", 4, 4
)
df2 <- df_dummy %>%
filter(label != "C")
myColors <- viridis::viridis_pal(option = "H")(3)
names(myColors) <- factor(df2$label)
dummy_col_scale <- scale_color_manual(name = "Legend", values = myColors)
df2 %>%
ggplot(aes(x, y, color = label))
geom_point()
dummy_col_scale
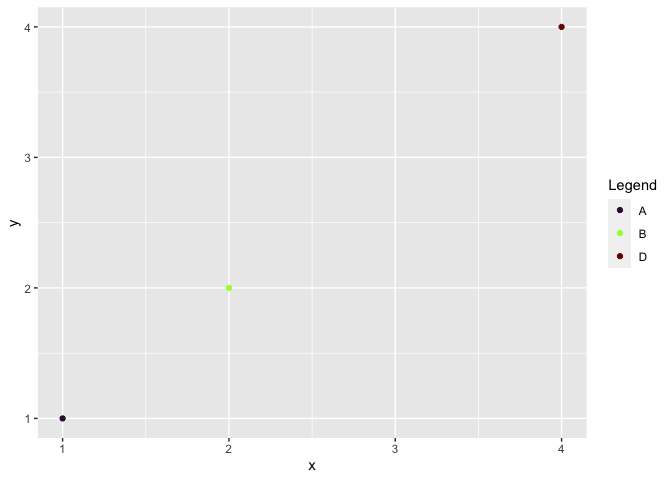
Another option is to set limits in dummy_col_scale, i.e.
df_dummy <- tribble(
~label, ~x, ~y,
"A", 1, 1,
"B", 2, 2,
"C", 3, 3,
"D", 4, 4
)
df2 <- df_dummy %>%
filter(label != "C")
myColors <- viridis::viridis_pal(option = "H")(4)
names(myColors) <- factor(df2$label)
dummy_col_scale <- scale_color_manual(name = "Legend", values = myColors,
limits = c("A", "B", "D"))
df2 %>%
ggplot(aes(x, y, color = label))
geom_point()
dummy_col_scale
CodePudding user response:
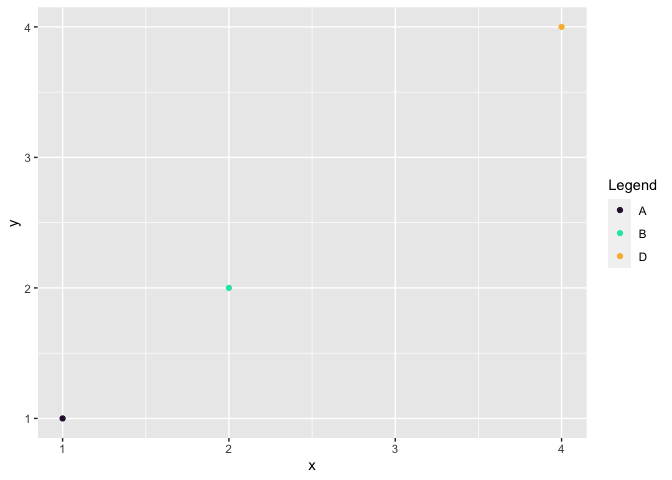
You may just simply remove C from myColors which is named vector, similar work with filter(label != "C").
myColors
A B C D
"#30123BFF" "#1AE4B6FF" "#FABA39FF" "#7A0403FF"
myColors[names(myColors) != "C"]
A B D
"#30123BFF" "#1AE4B6FF" "#7A0403FF"
Then, try
df_dummy %>%
filter(label != "C") %>%
ggplot(aes(x, y, color = label))
geom_point()
scale_color_manual(name = "Legend", values = myColors[names(myColors) != "C"])