I want to enter multiple variables into an R function, and I want to enter them all in the table1() function ==> something like this line tab<-table1(~ var1 var2 var3 ... varN|group, data=data)
library(table1)
dataset<-data.frame(ID=c(1,1,2,2,3,3,4,4),group=c("gp1","gp2","gp1","gp2","gp1","gp2","gp1","gp2"),
col1=c(0,1,1,1,0,1,1,0),col2=c(0,0,1,1,1,1,0,0),col3=c(1,0,1,0,1,1,1,0))
print.f <- function(data,var1,...,group){
tab<-table1(~ var1 ...|group, data=data)
tab
}
print.f(data,var1,var2,var3,group=group)
print.f(dataset,col1,col2,col3)
If I will have for example a dataset that contains more than 3 columns and I want to see their output, how could I enter all of them?
CodePudding user response:
Create the formula as a character vector, convert to formula class and run table1. In the examples we show several ways of creating the same output using print.f or using table1 directly.
print.f <- function(data, ..., group) {
v <- paste(c(...), collapse = " ")
if (!missing(group)) v <- paste(v, "|", group)
fo <- as.formula(paste("~", v))
table1(fo, data = data)
}
library(table1)
print.f(dataset, "col1", "col2", "col3", group = "group")
print.f(dataset, c("col1", "col2", "col3"), group = "group")
print.f(dataset, grep("col", names(dataset), value = TRUE), group = "group")
print.f(dataset, names(dataset)[2:4], group = "group")
print.f(dataset[-1], ".", group = "group")
print.f(dataset, ". - ID", group = "group")
table1(~ . | group, dataset[-1])
table1(~ . - ID | group, dataset)
CodePudding user response:
Here's a solution that manipulates language rather than strings. You and others might also find op_literal() useful in the future.
Solution
Helper: op_literal()
This helper function op_literal() actually manipulates the R language itself to repetitively use a binary operator like across many operands...even though a binary operator typically accepts only two operands. Calling op_literal(` `, w, x, y, z) will actually generate this expression here: w x y z.
# Helper function to arbitrarily repeat a binary operation (like ' ').
op_literal <- function(op, ...) {
# Capture the operator as a symbol.
op_sym <- rlang::ensym(op)
# Count the operands.
n_dots <- rlang::dots_n(...)
# Recursive case: a binary operator cannot handle this many arguments.
if(n_dots > 2) {
# Split off the final operand.
dots <- rlang::exprs(...)
dots_last <- dots[[n_dots]]
dots <- dots[-n_dots]
# Perform recursion for the remaining operands.
op_left <- rlang::inject(op_literal(
op = !!op_sym,
... = !!!dots
))
# Assemble recursive results into the full operation.
substitute(op(op_left, dots_last))
}
# Base case: the binary operator can handle 2(-) arguments.
else {
substitute(op(...))
}
}
Note
Since op_literal() generates an expression, you still need to evaluate it if you want the result:
op_exp <- op_literal(` `, 1, 2, 3, 4)
op_exp
#> 1 2 3 4
eval(op_exp)
#> [1] 10
Custom Function: print.f()
Next, this custom print.f() then leverages op_literal() to assemble the formula:
# Your custom 'print.f()' function.
print.f <- function(data, var1, ..., group) {
# Capture the core variables as symbols.
group_var <- rlang::ensym(group)
other_vars <- rlang::ensym(var1)
# Count the additional variables.
n_dots <- rlang::dots_n(...)
# Append those other variables if they exist.
if(n_dots > 0) {
other_vars <- rlang::inject(op_literal(op = ` `, !!other_vars, ...))
}
# Assemble the formula.
formula_exp <- rlang::inject(~ !!other_vars | !!group_var)
# Generate the table according to that formula.
table1::table1(
formula_exp,
data = data
)
}
Result
Given your dataset reproduced here
dataset <- data.frame(
ID = c(1, 1, 2, 2, 3, 3, 4, 4),
group = c("gp1", "gp2", "gp1", "gp2", "gp1", "gp2", "gp1", "gp2"),
col1 = c(0, 1, 1, 1, 0, 1, 1, 0),
col2 = c(0, 0, 1, 1, 1, 1, 0, 0),
col3 = c(1, 0, 1, 0, 1, 1, 1, 0)
)
your call to print.f()
print.f(dataset, col1, col2, col3, group = group)
should yield the following visualization:
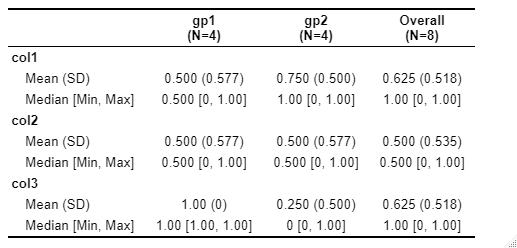
Note
As it stands, you have defined the group parameter at the end of your function header. This means that if you try calling print.f() like so
print.f(data = dataset, var = col1, col2, col3, group)
then you will get an error: without the group = specification, that final variable gets lumped together with col2 and col3, all under the ... umbrella. This will generate a bad formula:
~ col1 col2 col3 group |
To avoid the pain of having to type out group = every time, you can simply relocate it before the ..., like so:
print.f <- function(data, group, var1, ...) {
# ^^^^^
Once you've done so, the following call will work as you intended:
print.f(dataset, group, col1, col2, col3)

