I have a dataset with 40 sensors with odd names (e.g. A_B_Loc_1). I need to convert these data to long format to plot them. I need to split the names so I know the sensor name (e.g. from A_B_Loc_1, name=AB) and the sensor location (e.g. from A_B_Loc_1, location=1).
require(dplyr)
require(janitor)
require(tidyfast)
require(tidyr)
df<-data.frame(time=c("2021-02-27 22:06:20","2021-02-27 23:06:20"),A_Loc_1=c(500,600),A_Loc_2=c(500,600),A_B_Loc_1=c(500,600),A_B_Loc_2=c(500,600),B_Loc_1=c(500,600),B_3=c(500,600))
It's about 50 million rows, so it's very slow:
EDIT: Ouch! Some names don't have "Loc" (e.g. B_3 is sensor B, location 3).
#Pivoting it:
df %>%
tidyfast::dt_pivot_longer( #tidyfast package uses data.table instead of tidyr, so much faster
cols = -time,
names_to = "name",
values_to = "value"
) %>% drop_na()->df
#Split the names
df %>%
separate(name,
into = c("sensor", "location"),
sep = "(?=[0-9])"
) %>%
mutate(sensor=janitor::make_clean_names(sensor, case = "big_camel"))
Can this be sped up? A left join with a lookup table that adds columns based on sensor names?
CodePudding user response:
library(data.table)
setDT(df)
dt <- melt(df, id.vars = c("time"))
dt[, c("name", "location") := tstrsplit(str_replace_all(variable, "_", ""), "Loc")]
dt
# time variable value name location
# 1: 2021-02-27 22:06:20 A_Loc_1 500 A 1
# 2: 2021-02-27 23:06:20 A_Loc_1 600 A 1
# 3: 2021-02-27 22:06:20 A_Loc_2 500 A 2
# 4: 2021-02-27 23:06:20 A_Loc_2 600 A 2
# 5: 2021-02-27 22:06:20 A_B_Loc_1 500 AB 1
# 6: 2021-02-27 23:06:20 A_B_Loc_1 600 AB 1
# 7: 2021-02-27 22:06:20 A_B_Loc_2 500 AB 2
# 8: 2021-02-27 23:06:20 A_B_Loc_2 600 AB 2
# 9: 2021-02-27 22:06:20 B_Loc_1 500 B 1
# 10: 2021-02-27 23:06:20 B_Loc_1 600 B 1
Edit: OP mentions that Loc is not always present, so we split on the last underscore to get the number. Then we clean the name in the second step to remove the underscores and - if present - "Loc"
dt <- melt(df, id.vars = c("time"))
dt[, c("name", "location") := tstrsplit(variable, "_(?!.*_)", perl = T)]
dt[, name := str_replace_all(name, "_|Loc", "")]
CodePudding user response:
We experimented with a few approaches to splitting columns by regular expressions. separate was very slow, but the fastest seems to be stringr::str_split(..., simplify=TRUE) to make new columns (for a tibble):
require(dplyr)
require(janitor)
require(tidyr)
require(stringr)
df <-
data.frame(
time = c("2021-02-27 22:06:20", "2021-02-27 23:06:20"),
A_Loc_1 = c(500, 600),
A_Loc_2 = c(500, 600),
A_B_Loc_1 = c(500, 600),
A_B_Loc_2 = c(500, 600),
B_Loc_1 = c(500, 600)
)
df1 <- df %>%
# Suggestion from above about cleaning names first?
clean_names(case = "big_camel") %>%
tidyfast::dt_pivot_longer(
cols = -Time,
names_to = "name",
values_to = "value") %>%
drop_na() %>%
as_tibble
df1[c("sensor", "location")] <-
str_split(df1$name, "Loc", simplify = TRUE)
This is presuming your biggest time guzzler is the separating columns part!
Edit
There are at least four ways of splitting, and depending on the complexity of the split it might be quicker to use other methods (such as data.table::tstrsplit), but some of these would require a consistent 'split' across all rows:
library(tidyverse)
library(data.table)
# a sample of 100,000 pivoted rows
n <- 1e5
df <- data.frame(condition = c(rep("ABLoc1", times = n),
rep("ABLoc2", times = n),
rep("ACLoc1", times = n),
rep("ACLoc2", times = n),
rep("AALoc4", times = n)))
(speeds <- bench::mark(
separate = {
df_sep <- df %>%
separate(condition,sep = "Loc", into = c("part1", "part2"), remove = FALSE)
},
dt = {
df_dt <- data.table::data.table(df)
df_dt <-
df_dt[, c("part1" , "part2") := tstrsplit(condition, split = "Loc", fixed = TRUE)]
},
stringr = {
df_str <- df
df_str[c("part1", "part2")] <- str_split(df_str$condition, "Loc", simplify = TRUE)
},
gsub = {
df_vec <- df
df_vec$part1 <- gsub("(^.*)Loc.*", "\\1", df$condition)
df_vec$part2 <- gsub(".*Loc(.*$)", "\\1", df$condition)
},
iterations = 10,
check = FALSE
))
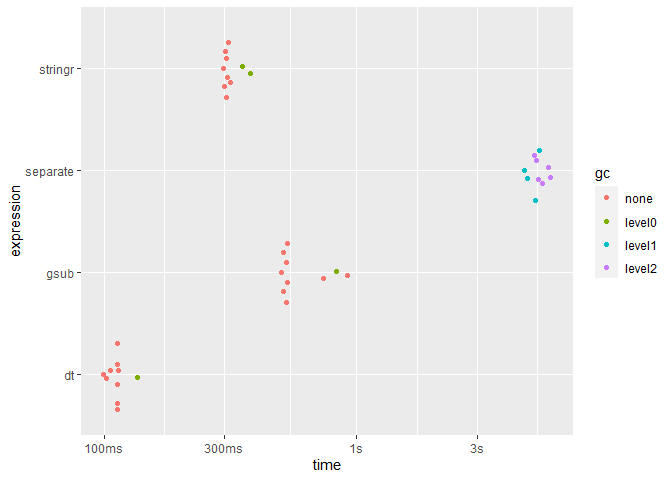
#> # A tibble: 4 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 separate 4.63s 5.19s 0.191 3.89GB 4.25
#> 2 dt 99.44ms 112.32ms 8.95 28.91MB 0.895
#> 3 stringr 296.11ms 306.5ms 3.16 59.53MB 0.632
#> 4 gsub 502.85ms 528.69ms 1.63 7.63MB 0.163
plot(speeds, type = "beeswarm")
Plotting speeds of each approach (to iterate over 100,000 rows):
Created on 2021-12-08 by the reprex package (v2.0.1)
