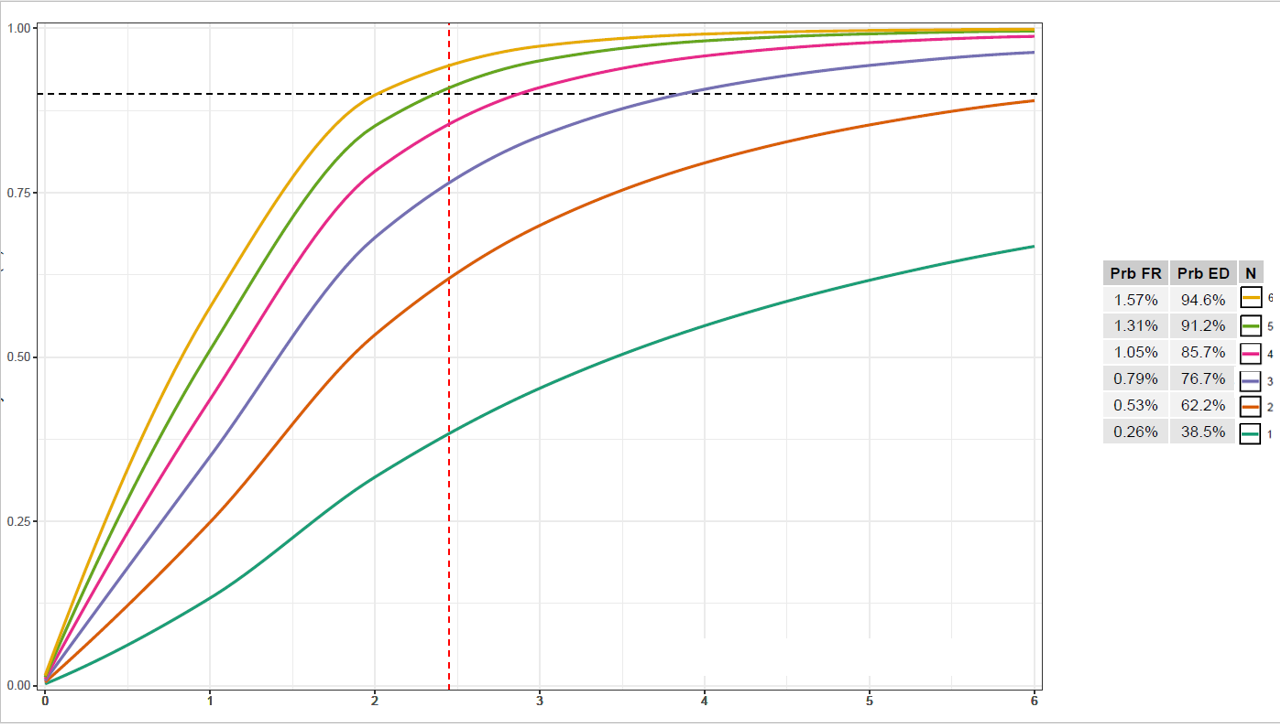
I was wondering if anyone knows a way to combine a table and ggplot legend so that the legend appears as a column in the table as shown in the image. Sorry if this has been asked before but I haven't been able to find a way to do this.
Edit: attached is code to produce the output below (minus the legend/table combination, which I am trying to produce, as I stitched that together in Powerpoint)
library(ggplot2)
library(gridExtra)
library(dplyr)
library(formattable)
library(signal)
#dataset for ggplot
full.data <- structure(list(error = c(0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4,
5, 6, 0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4,
5, 6, 0, 1, 2, 3, 4, 5, 6), prob.ed.n = c(0, 0, 0.2, 0.5, 0.8,
1, 1, 0, 0, 0.3, 0.7, 1, 1, 1, 0, 0.1, 0.4, 0.9, 1, 1, 1, 0,
0.1, 0.5, 0.9, 1, 1, 1, 0, 0.1, 0.6, 1, 1, 1, 1, 0, 0.1, 0.6,
1, 1, 1, 1), N = c(1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2,
3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 5, 5, 5, 5, 5, 5, 5,
6, 6, 6, 6, 6, 6, 6)), row.names = c(NA, -42L), class = "data.frame")
#summary table
summary.table <- structure(list(prob.fr = c("1.62%", "1.35%", "1.09%", "0.81%", "0.54%", "0.27%"), prob.ed.n = c("87.4%", "82.2%", "74.8%", "64.4%", "49.8%", "29.2%"), N = c(6, 5, 4, 3, 2, 1)), row.names = c(NA,
-6L), class = "data.frame")
#table object to beincluded with ggplot
table <- tableGrob(summary.table %>%
rename(
`Prb FR` = prob.fr,
`Prb ED` = prob.ed.n,
),
rows = NULL)
#plot
plot <- ggplot(full.data, aes(x = error, y = prob.ed.n, group = N, colour = as.factor(N)))
geom_vline(xintercept = 2.45, colour = "red", linetype = "dashed")
geom_hline(yintercept = 0.9, linetype = "dashed")
geom_line(data = full.data %>%
group_by(N) %>%
do({
tibble(error = seq(min(.$error), max(.$error),length.out=100),
prob.ed.n = pchip(.$error, .$prob.ed.n, error))
}),
size = 1)
scale_x_continuous(labels = full.data$error, breaks = full.data$error, expand = c(0, 0.05))
scale_y_continuous(expand = expansion(add = c(0.01, 0.01)))
scale_color_brewer(palette = "Dark2")
guides(color = guide_legend(reverse=TRUE, nrow = 1))
theme_bw()
theme(legend.key = element_rect(fill = "white", colour = "black"),
legend.direction= "horizontal",
legend.position=c(0.8,0.05)
)
#arrange plot and grid side-by-side
grid.arrange(plot, table, nrow = 1, widths = c(4,1))
CodePudding user response:
This is an interesting problem. The short answer: Yes it's possible. But it's currently very ugly. Although I will probably never really need that visualisation, I think that I'll put a bounty on that, once possible.
As per r2evans' comment, the alignment of the legend with the table is probably the most difficult bit. Here the ugly hack, using ggpubr for the table, and cowplot for the stitching.
Another problem arises from the legend key spacing for vertical legends. This is still a rather unresolved issue for other keys than polygons, to my knowledge. The associated GitHub issue is closed, but there might be new ones out there. From a brief glance at the draw_key function used in ggplot2:::GeomLine, it is not clear to me where to change the distance between the different keys. I think I'll post this as a new question soon, but today it's too late.
Some other relevant comments in the code.
library(tidyverse)
library(ggpubr)
library(cowplot)
#>
#> Attaching package: 'cowplot'
#> The following object is masked from 'package:ggpubr':
#>
#> get_legend
# dataset for ggplot
full.data <- structure(list(error = c(
0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4,
5, 6, 0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4,
5, 6, 0, 1, 2, 3, 4, 5, 6
), prob.ed.n = c(
0, 0, 0.2, 0.5, 0.8,
1, 1, 0, 0, 0.3, 0.7, 1, 1, 1, 0, 0.1, 0.4, 0.9, 1, 1, 1, 0,
0.1, 0.5, 0.9, 1, 1, 1, 0, 0.1, 0.6, 1, 1, 1, 1, 0, 0.1, 0.6,
1, 1, 1, 1
), N = c(
1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2,
3, 3, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 4, 4, 5, 5, 5, 5, 5, 5, 5,
6, 6, 6, 6, 6, 6, 6
)), row.names = c(NA, -42L), class = "data.frame")
summary.table <-
structure(list(
prob.fr = c("1.62%", "1.35%", "1.09%", "0.81%", "0.54%", "0.27%"),
prob.ed.n = c("87.4%", "82.2%", "74.8%", "64.4%", "49.8%", "29.2%"),
N = c(6, 5, 4, 3, 2, 1)
), row.names = c(NA, -6L), class = "data.frame")
## Hack number zero - create some space for the new legend
## this is not great, as not automated
spacer <- paste(rep(" ", 7), collapse = "")
my_table <-
summary.table %>%
mutate(N = paste(spacer, N))
p1 <-
ggplot(full.data, aes(x = error, y = prob.ed.n, group = N, colour = as.factor(N)))
geom_vline(xintercept = 2.45, colour = "red", linetype = "dashed")
geom_hline(yintercept = 0.9, linetype = "dashed")
geom_line(
data = full.data %>%
group_by(N) %>%
do({
tibble(
error = seq(min(.$error), max(.$error), length.out = 100),
prob.ed.n = signal::pchip(.$error, .$prob.ed.n, error)
)
}),
size = 1
)
## hack 1 - remove the legend labels. You have them in the table already.
scale_color_brewer(NULL, palette = "Dark2", labels = rep("", 6))
## remove all the legend specs! I've also removed the not so important reverse scale
## I have to remove fill and color, otherwise the hack becomes too evident
theme(
legend.key = element_rect(fill = NA, colour = NA),
legend.key.height = unit(.27, "in"),
legend.background = element_blank()
)
## create the plot elements
p_leg <- cowplot::get_legend(p1)
p2 <- ggtexttable(my_table, rows = NULL)
## we don't want the legend twice
p <- p1 theme(legend.position = "none")
## the positioning is the horrible bit and totally lacks automation.
ggdraw(p, xlim = c(0, 1.7))
draw_plot(p2, x = .8)
draw_plot(p_leg, x = .97, y = 0.98, vjust = 1)
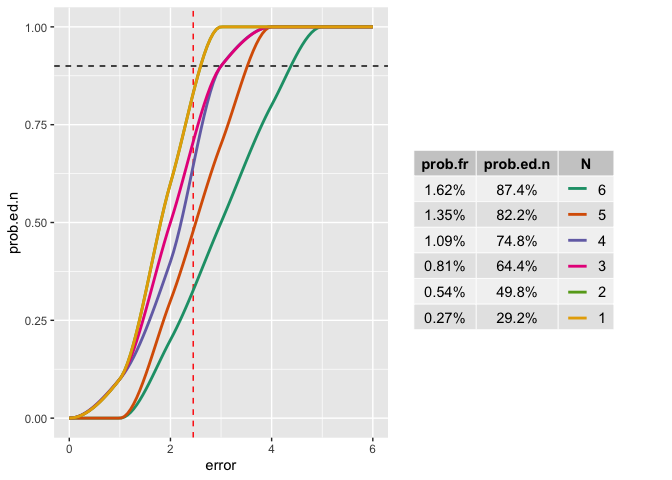
Created on 2021-12-29 by the reprex package (v2.0.1)