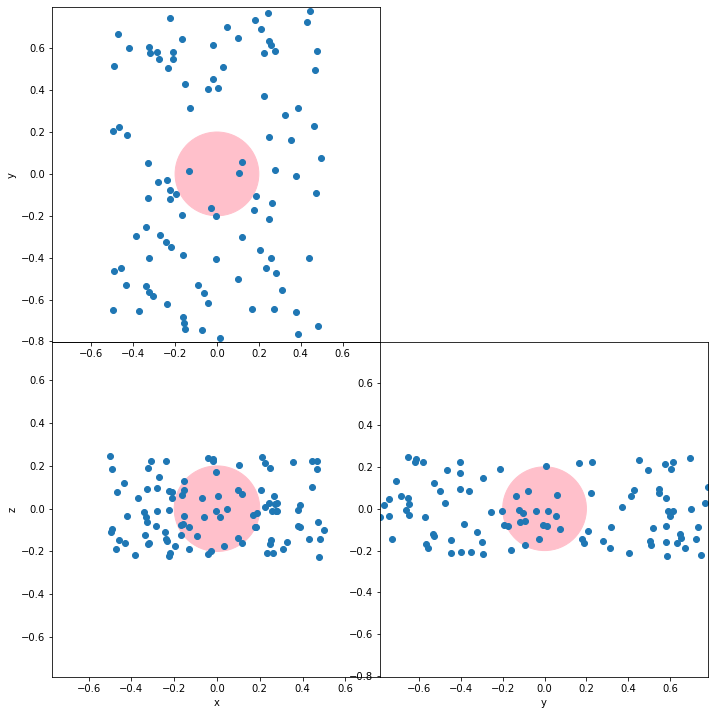
I'm trying to plot 3D data in 2D using orthographic projection. Here is partially what I'm looking for:
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
fig = plt.figure(figsize=(10,10),facecolor='white')
axs = [fig.add_subplot(223)]
axs.append(fig.add_subplot(224))#,sharey=axs[0]))
axs.append(fig.add_subplot(221))#,sharex=axs[0]))
rng = np.random.default_rng(12345)
values = rng.random((100,3))-.5
values[:,1] = 1.6*values[:,1]
values[:,2] = .5*values[:,2]
for ax,axis in zip(axs,['y','x','z']):
axis1,axis2={'x':(1,2),'y':(0,2),'z':(0,1)}[axis]
ax.add_patch(plt.Circle([0,0], radius=.2, color='pink',zorder=-20))
ax.scatter(values[:,axis1],values[:,axis2])
axs[0].set_xlabel('x')
axs[2].set_ylabel('y')
axs[1].set_xlabel('y')
axs[0].set_ylabel('z')
fig.subplots_adjust(.08,.06,.99,.99,0,0)
plt.show()
There are some issues with this plot and the fixes I tried: I would need 'equal' aspect so that the circles are actually circle. I would also need the circles to be of the same size in each subplot. Finally, I would like the space to be optimized (i.e. with as little white space inside and between the subplots as possible).
I have tried sharing the axis between the subplots, then doing .axis('scaled') or .set_aspect('equal','box',share=True) for each axes, but the axis end up not being properly shared, and the circle in each subplot end up of different sizes. And while it crops the subplots to the data, it leaves a lot of space between the subplots. .axis('equal') or .set_aspect('equal','datalim',share=True) without axis shared leaves white space inside the subplots, and with shared axis, it leaves out some data.
Any way to make it work? And it would be perfect if it can work on matplotlib 3.4.3.
CodePudding user response:
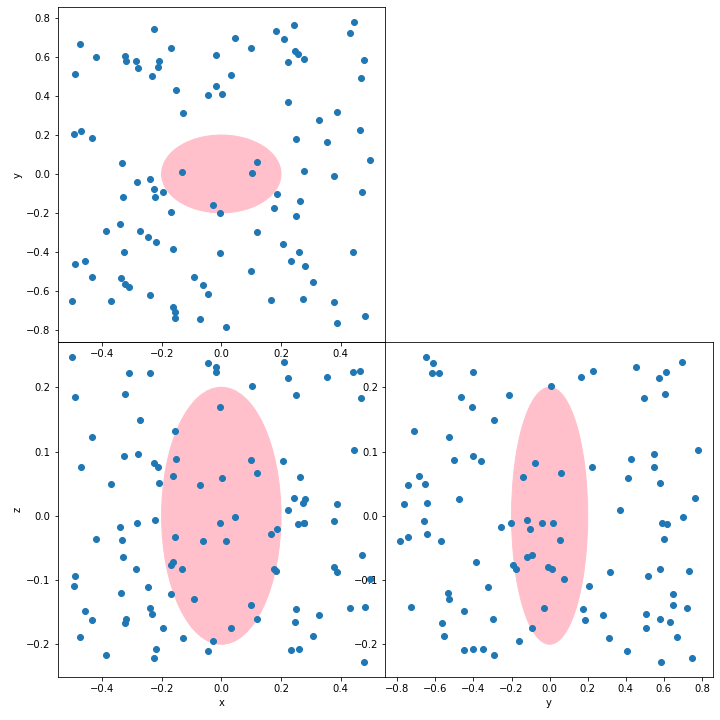
You can use a common xlim, ylim for your subplots and set your equal ratio with ax.set_aspect(aspect='equal', adjustable='datalim'):
See full code below:
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
fig = plt.figure(figsize=(10,10),facecolor='white')
axs = [fig.add_subplot(223)]
axs.append(fig.add_subplot(224))#,sharey=axs[0]))
axs.append(fig.add_subplot(221))#,sharex=axs[0]))
rng = np.random.default_rng(12345)
values = rng.random((100,3))-.5
values[:,1] = 1.6*values[:,1]
values[:,2] = .5*values[:,2]
for ax,axis in zip(axs,['y','x','z']):
axis1,axis2={'x':(1,2),'y':(0,2),'z':(0,1)}[axis]
ax.add_patch(plt.Circle([0,0], radius=.2, color='pink',zorder=-20))
ax.scatter(values[:,axis1],values[:,axis2])
ax.set_xlim([np.amin(values),np.amax(values)])
ax.set_ylim([np.amin(values),np.amax(values)])
ax.set_aspect('equal', adjustable='datalim')
axs[0].set_xlabel('x')
axs[2].set_ylabel('y')
axs[1].set_xlabel('y')
axs[0].set_ylabel('z')
fig.subplots_adjust(.08,.06,.99,.99,0,0)
plt.show()
CodePudding user response:
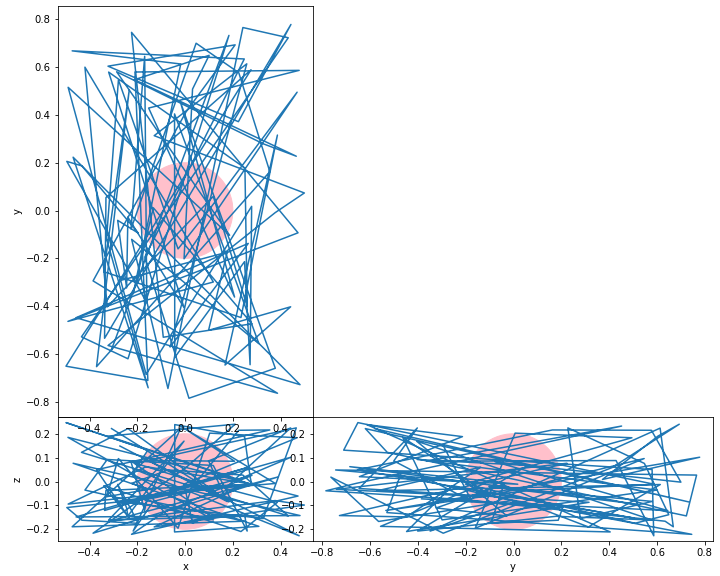
I made it work using gridspec (I changed scatter for plot to visually make sure no data gets left out). It requires some tweaking of the figsize to really minimize the white space within the axes. Thank you to @jylls for the intermediate solution.
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
%matplotlib inline
rng = np.random.default_rng(12345)
values = rng.random((100,3))-.5
values[:,1] = 1.6*values[:,1]
values[:,2] = .5*values[:,2]
fig = plt.figure(figsize=(10,8),facecolor='white')
ranges = np.ptp(values,axis=0)
gs = GridSpec(2, 2, None,.08,.06,.99,.99,0,0, width_ratios=[ranges[0], ranges[1]], height_ratios=[ranges[1], ranges[2]])
axs = [fig.add_subplot(gs[2])]
axs.append(fig.add_subplot(gs[3]))#,sharey=axs[0]))
axs.append(fig.add_subplot(gs[0]))#,sharex=axs[0]))
for ax,axis in zip(axs,['y','x','z']):
axis1,axis2={'x':(1,2),'y':(0,2),'z':(0,1)}[axis]
ax.add_patch(plt.Circle([0,0], radius=.2, color='pink',zorder=-20))
ax.plot(values[:,axis1],values[:,axis2])
ax.set_aspect('equal', adjustable='datalim')
axs[0].set_xlabel('x')
axs[2].set_ylabel('y')
axs[1].set_xlabel('y')
axs[0].set_ylabel('z')
plt.show()