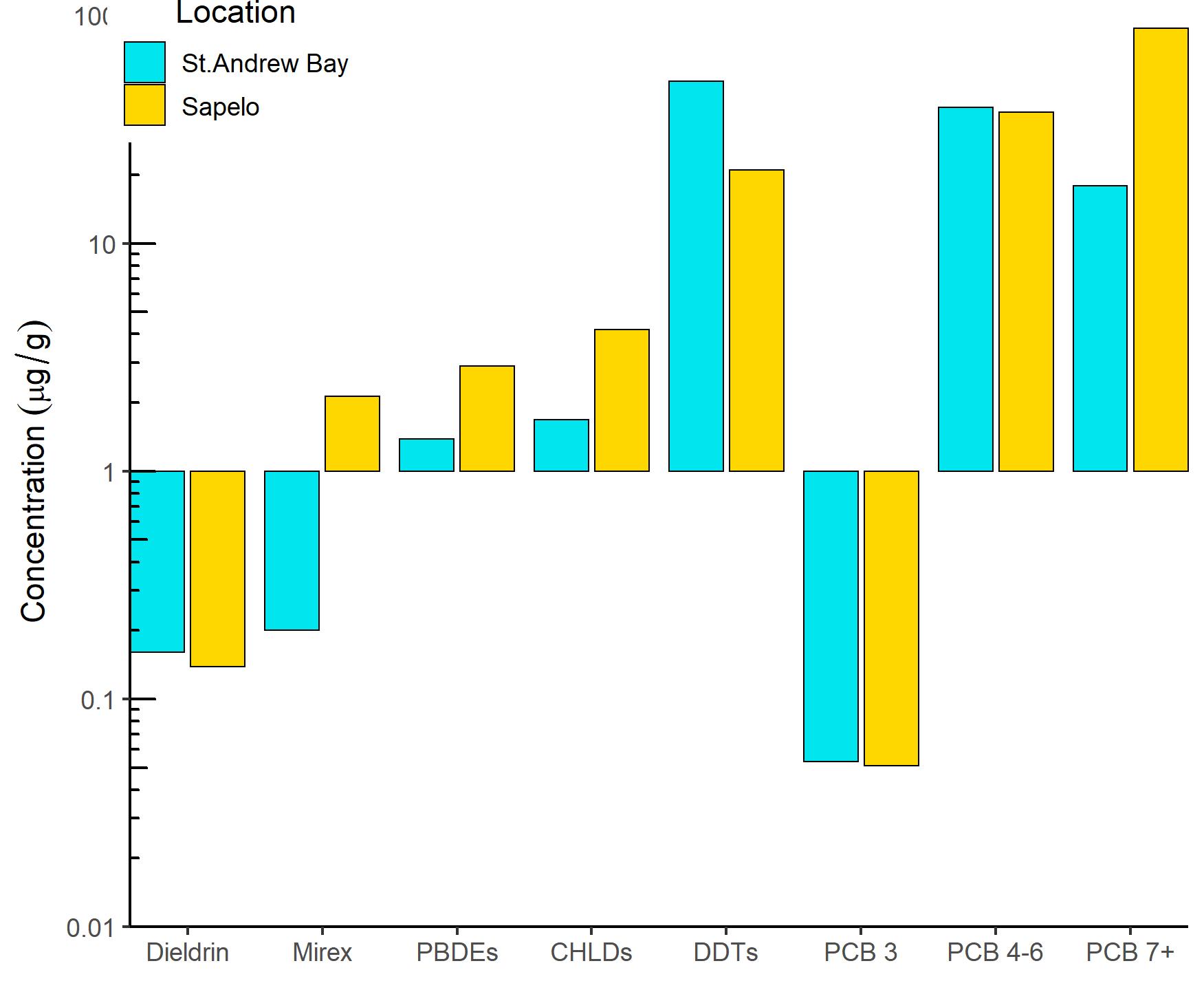
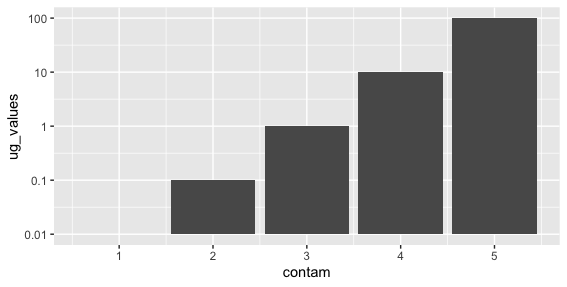
I am trying to make a grouped bar plot comparing the concentration of contaminants (in ug/g) from 2 different locations site comparison figure, with the y axis in log scale. I have some values that are below one but greater than zero. The y axis is centering my bars at y=1, making some of the bars look negative. Is there to make my bars start at 0.01 instead of 1?
I tried coord_cartesian( ylim=c(0.01,100), expand = FALSE ) but that didn't do it. I also tried to use coor_trans(y='log10') to log transform y axis instead of scale_y_continuous(trans='log10') but was getting the error message "Transformation introduced infinite values in y-axis" even though I have no zero values.
Any help would be much appreciated, Thank you.
my code is below:
malcomp2 %>%
ggplot(aes(x= contam, y= ug_values, fill= Location))
geom_col(data= malcomp2,
mapping = aes(x= contam, y= ug_values, fill = Location),
position = position_dodge(.9), #makes the bars grouped
stat_identity(),
colour = "black", #adds black lines around bars
width = 0.8,
size = 0.3)
ylab(expression(Concentration~(mu*g/g))) #adds mu character to y axis
coord_cartesian( ylim=c(0.01,100), expand = FALSE ) #force bars to start at 0
theme_classic() #get rid of grey grid
scale_y_continuous(trans='log10', #change to log scale
labels = c(0.01,0.1,1,10,100)) #change axis to not sci notation
annotation_logticks(sides = 'l') #add log ticks to y axis only
scale_x_discrete(name = NULL, #no x axis label
limits = c('Dieldrin','Mirex','PBDEs','CHLDs','DDTs','PCBtri','PCBquad',
'PCBhept'), #changes order of x axis
labels = c('Dieldrin','Mirex','PBDEs','CHLDs','DDTs','PCB 3','PCB 4-6','PCB7 '))
scale_fill_manual("Location", #rename legend
values = c('turquoise2','gold'), #change colors
labels = c( 'St.Andrew Bay', 'Sapelo')) #change names on legend
theme(legend.title = NULL,
legend.key.size = unit(15, "pt"),
legend.position = c(0.10,0.95)) #places legend in upper left corner
CodePudding user response:
One hack would be to just scale your data so baseline is at 1. People have asked this question before on SO, and it seems like a more satisfactory approach might be to use geom_rect instead, like here: