I want to subset rows based on different column conditions across several columns. For example, in the attached picture, I want my script to remove all rows that meet the following conditions:
In any of the columns (One through Five), remove rows where no valid entry is made in any of the five columns (valid entries are: poor, good, very_good, excellent). In essence, remove rows with invalid entries (invalid entries are: "NULL", "''" or contains "@")
In this example, only Chris will be excluded and others will be retained since they contain at least one valid entry across the 5 columns.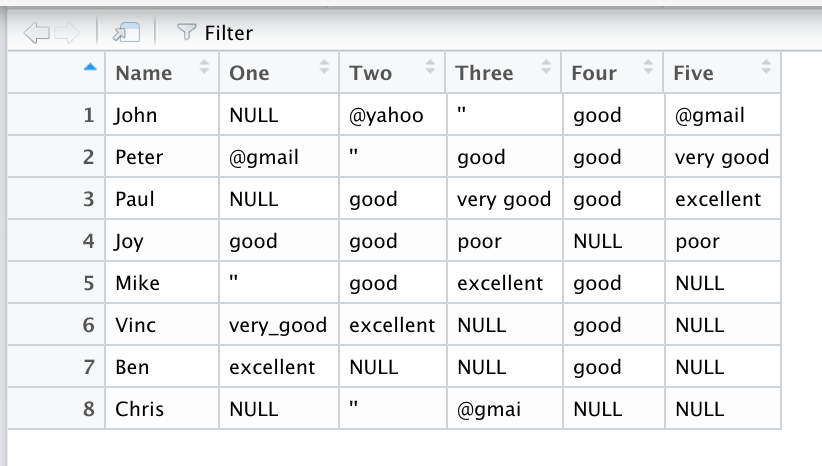
Data:
df <-
tibble(
Name = c("John", "Peter", "Paul", "Joy", "Mike", "Vinc", "Ben", "Chris"),
One = c("NULL", "@gmail", "NULL", "good", "''", "very_good", "excellent", "NULL"),
Two = c("@yahoo", "''", "good", "good", "good", "excellent", "NULL", "''"),
Three = c("''", "good", "very good", "poor", "excellent", "NULL", "NULL", "@gmai"),
Four = c("good", "good", "good", "NULL", "good", "good", "good", "NULL"),
Five = c("@gmail", "very good", "excellent", "poor", "NULL", "NULL", "NULL", "NULL")
)
CodePudding user response:
You may use dplyr::if_any here -
library(dplyr)
valid_entry <- c("poor", "good", "very_good", "excellent")
df %>% filter(if_any(One:Five, ~.x %in% valid_entry))
CodePudding user response:
EDIT:
To filter rows where any columns from One to Five contain 'invalid' values:
library(dplyr)
library(stringr)
df %>%
filter(if_any(One:Five,
~!str_detect(., paste0(c("poor", "good", "very_good", "excellent"), collapse = "|"))))
# A tibble: 6 × 6
Name One Two Three Four Five
<chr> <chr> <chr> <chr> <chr> <chr>
1 John NULL @yahoo '' good @gmail
2 Peter @gmail '' good good very good
3 Paul NULL good very good good excellent
4 Mike '' good excellent good NULL
5 Ben excellent NULL NULL good NULL
6 Chris NULL '' @gmai NULL NULL
To filter rows where all columns from One to Five contain 'invalid' values:
library(dplyr)
library(stringr)
df %>%
filter(if_all(One:Five,
~!str_detect(., paste0(c("poor", "good", "very_good", "excellent"), collapse = "|"))))
# A tibble: 1 × 6
Name One Two Three Four Five
<chr> <chr> <chr> <chr> <chr> <chr>
1 Chris NULL '' @gmai NULL NULL
Data:
df <-
tibble(
Name = c("John", "Peter", "Paul", "Joy", "Mike", "Vinc", "Ben", "Chris"),
One = c("NULL", "@gmail", "NULL", "good", "''", "very_good", "excellent", "NULL"),
Two = c("@yahoo", "''", "good", "good", "good", "excellent", "NULL", "''"),
Three = c("''", "good", "very good", "poor", "excellent", "NULL", "NULL", "@gmai"),
Four = c("good", "good", "good", "NULL", "good", "good", "good", "NULL"),
Five = c("@gmail", "very good", "excellent", "poor", "NULL", "NULL", "NULL", "NULL")
)
CodePudding user response:
You can try this:
# sample data
df <- data.frame(
Name = c("John", "Peter", "Chris"),
One = c("NULL", "@gmail", "NULL"),
Two = c("@yahoo", "", "@gmai"),
Three = c("very good", "good", "NULL")
)
Function that checks if a row is valid or not:
isInvalid <- function(row) {
row <- row[-1] # ignore Name
ats <- length(grep("@", row)) # count the number of cells with "@"
invalids <- c("NULL", "") # list of error values
invs <- length(which(row %in% invalids)) # count nr of error values
(ats invs) == length(row) # if 'nr of ats' 'nr of error values' is equal to the nr of cells -> invalid row
}
Call function for each row in the dataframe:
invalidrows <- apply(df, MARGIN = 1, FUN = isInvalid)
# results in FALSE FALSE TRUE
Extract the invalid rows from the original dataframe:
invalid <- df[invalidrows,]
# returns the 'Chris' row
