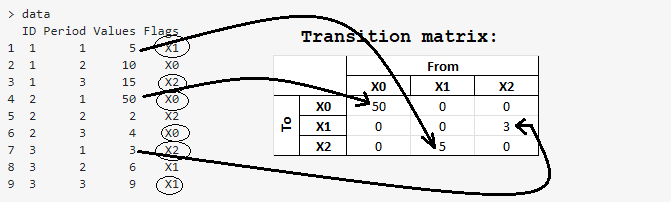
In the below reproducible code, the custom balTransit() function correctly populates a values transition table using a for-loop, while the custom balTransit_1() function is supposed to do the same using lapply() but it doesn't work. What am I doing wrong in my implementation of lapply()? Run the code and you'll see results of:
balTransit (correct results):
> test
X1 X0 X2
X1 0 0 3
X0 0 50 0
X2 5 0 0
balTransit_1 (incorrect, all 0's):
> test_1
X1 X0 X2
X1 0 0 0
X0 0 0 0
X2 0 0 0
Enhanced explanation:
My main objective here is to learn how to use the apply() family of functions, for their perceived benefits. I’ve been going through simple tutorials. A secondary objective is the generation of a transition matrix from a base data frame. Once I figure this out with lapply() (or another apply() function that is most suitable), I’m going to run the various options (for-loop(), data.table(), lapply(), etc.) against the actual data set of 2.5m rows for speed testing.
What I’m doing is creating a transition matrix (technically here a data frame) showing the flow of values (balances) from one “Flags” category to another “Flags” category, over the periods specified by the user. So, in my “for-loop” reproducible example which works correctly, the user has specified a “From” period of 1 and a “To” period of 3. The transition matrix is then generated as shown in the image now posted at the bottom.
CodePudding user response:
To make your lapply code work just replace <- with <<-:
DF_1[row, column] <<- DF_1[row, column] val
Please see ?assignOps for more info.
However, again I wouldn't recommend lapply in this case (<<- should be avoided in general)
Here is a data.table approach:
library(data.table)
DT <- setDT(data.frame(
ID = c(1,1,1,2,2,2,3,3,3),
Period = c(1, 2, 3, 1, 2, 3, 1, 2, 3),
Values = c(5, 10, 15, 50, 2, 4, 3, 6, 9),
Flags = c("X1","X0","X2","X0","X2","X0", "X2","X1","X1")
))
unique_flags <- unique(DT$Flags)
all_flags <- setDT(expand.grid(list(first_flag = unique_flags, last_flag = unique_flags)))
resultDT <- dcast(
data = DT[, .(first_flag = first(Flags), last_flag = last(Flags), first_value = first(Values)), by = ID][
all_flags, on = c("first_flag", "last_flag")],
last_flag ~ first_flag,
fun.aggregate = sum,
value.var = "first_value"
)
for (col_i in seq_len(ncol(resultDT))){
set(resultDT, which(is.na(resultDT[[col_i]])), col_i, 0)
}
print(resultDT)
Result:
last_flag X0 X1 X2
1: X0 50 0 0
2: X1 0 0 3
3: X2 0 5 0
# step by step ------------------------------------------------------------
library(data.table)
DT <- setDT(data.frame(
ID = c(1,1,1,2,2,2,3,3,3,4,4,4),
Period = c(1, 2, 3, 1, 2, 3, 1, 2, 3, 1, 2, 3),
Values = c(5, 10, 15, 50, 2, 4, 3, 6, 9, 3, 6, 9),
Flags = c("X1","X0","X2","X0","X2","X0", "X2","X1","X1", "X2","X1","X1")
))
unique_flags <- unique(DT$Flags)
all_flags <- setDT(expand.grid(list(first_flag = unique_flags, last_flag = unique_flags)))
resultDT <- DT[, .(first_flag = first(Flags), last_flag = last(Flags), first_value = first(Values)), by = ID] # find relevant flags
resultDT <- resultDT[all_flags, on = c("first_flag", "last_flag")] # merge all combinations
resultDT <- dcast(resultDT, last_flag ~ first_flag, fun.aggregate = sum, value.var = "first_value") # dcast
for (col_i in seq_len(ncol(resultDT))){
set(resultDT, which(is.na(resultDT[[col_i]])), col_i, 0)
}
print(resultDT)