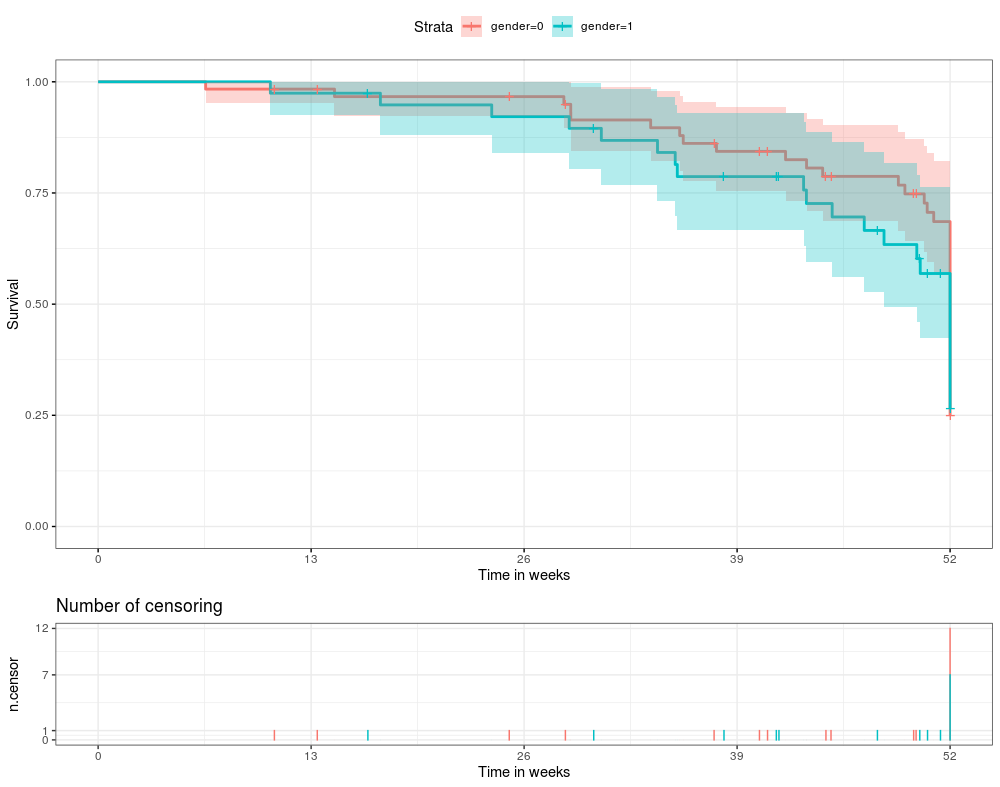
The ggsurvplot function from the package survminer allows users to draw Kaplen-Meier curves. It also allows us to create a table beneath the plot with the number of censored observations by configuring the ncensor.plot value. My question is: how does one change the y-axis of the censored table beneath the plot? The reason is I have a study termination at for example 52 weeks, and therefore the amount of censored observations is high, skewing the table completely. See the example below.
#Create a random dataset with gender, event, time
set.seed(100)
time <- rnorm(n = 100, mean = 52, sd = 20)
set.seed(150)
gender <- sample(x = c(0, 1), prob = c(.6, .4), size = 100, replace = TRUE)
set.seed(100)
event <- sample(x = c(0, 1), prob = c(.4, .6), size = 100, replace = TRUE)
dat <- data.frame(time, gender, event)
#Censor time after 52 weeks
dat <- dat %>% mutate(time_adjusted = if_else(time > 52, 52, time))
#KM-estimator
survObject <- Surv(time = dat$time_adjusted, event = dat$event)
km.model <- survfit(survObject ~ gender, data = dat)
summary(km.model)
Call: survfit(formula = survObject ~ gender, data = dat)
gender=0
time n.risk n.event survival std.err lower 95% CI upper 95% CI
6.56149 61 1 0.983607 0.0162585 0.952251 1.000000
14.42688 58 1 0.966648 0.0231936 0.922242 1.000000
28.42634 56 1 0.949386 0.0284876 0.895162 1.000000
28.84541 54 1 0.931805 0.0329415 0.869427 0.998658
28.84858 53 1 0.914224 0.0367130 0.845027 0.989087
33.72372 52 1 0.896643 0.0399957 0.821581 0.978562
35.49481 51 1 0.879061 0.0429020 0.798871 0.967301
35.71242 50 1 0.861480 0.0455040 0.776755 0.955446
37.73950 48 1 0.843533 0.0479650 0.754572 0.942981
41.95615 45 1 0.824787 0.0504291 0.731641 0.929793
43.23820 44 1 0.806042 0.0526518 0.709180 0.916135
44.22292 43 1 0.787297 0.0546624 0.687131 0.902065
48.84190 40 1 0.767615 0.0567288 0.664106 0.887257
49.24141 39 1 0.747932 0.0585893 0.641480 0.872050
50.42166 36 1 0.727156 0.0605334 0.617686 0.856027
50.60166 35 1 0.706380 0.0622672 0.594300 0.839599
51.00008 34 1 0.685605 0.0638078 0.571287 0.822798
52.00000 33 21 0.249311 0.0619235 0.153222 0.405659
gender=1
time n.risk n.event survival std.err lower 95% CI upper 95% CI
10.5119 39 1 0.974359 0.0253102 0.925994 1.000000
17.2280 37 1 0.948025 0.0357936 0.880404 1.000000
24.0235 36 1 0.921691 0.0434190 0.840402 1.000000
28.7516 35 1 0.895357 0.0495247 0.803367 0.997880
30.7129 33 1 0.868225 0.0549558 0.766927 0.982903
34.1409 32 1 0.841093 0.0595606 0.732095 0.966319
35.2230 31 1 0.813961 0.0635192 0.698519 0.948482
35.3501 30 1 0.786829 0.0669463 0.665973 0.929616
43.0588 26 1 0.756566 0.0708822 0.629649 0.909066
43.2310 25 1 0.726303 0.0742265 0.594466 0.887379
44.8028 24 1 0.696041 0.0770563 0.560275 0.864706
46.7601 23 1 0.665778 0.0794266 0.526965 0.841157
47.9673 21 1 0.634074 0.0817272 0.492524 0.816307
49.9674 20 1 0.602371 0.0835642 0.458966 0.790583
50.1777 18 1 0.568906 0.0853600 0.423959 0.763408
52.0000 15 8 0.265489 0.0834090 0.143425 0.491439
#KM-curves
ggsurvplot(km.model, data = dat, ggtheme = theme_bw(), conf.int = TRUE, risktable = "abs_pct", ncensor.plot = TRUE,
xlab = "Time in weeks", ylab = "Survival", xlim = c(0,52), break.x.by = 13)
CodePudding user response:
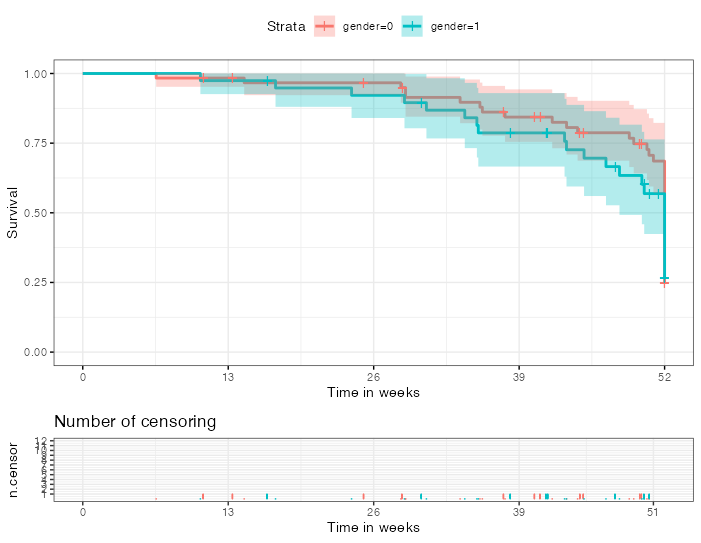
The object returned by ggsurvplot is a list which contains the main plot as an element called plot and the censored table plot as an element named ncensor.plot. Both of which are ggplot objects and can therefore be tweaked using default ggplot options. As an example in my code below I use scale_y_continuous to change the breaks of the y scale for the censored table plot:
library(survminer)
library(survival)
library(dplyr)
p <- ggsurvplot(km.model, data = dat, ggtheme = theme_bw(), conf.int = TRUE, risktable = "abs_pct", ncensor.plot = TRUE,
xlab = "Time in weeks", ylab = "Survival", xlim = c(0,52), break.x.by = 13)
p$ncensor.plot <- p$ncensor.plot scale_y_continuous(breaks = seq(12))
#> Scale for 'y' is already present. Adding another scale for 'y', which will
#> replace the existing scale.
p
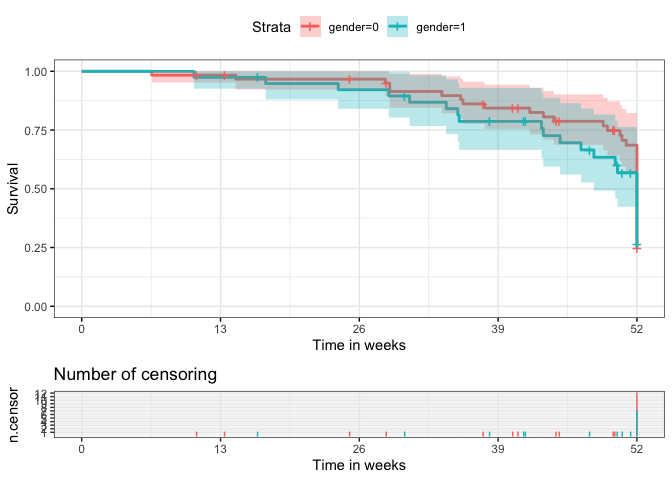
EDIT The limits of the x axis could be set using the limits argument, where I also have set the breaks according to ones set by ggsurvplot by default, except for the 52 which I replaced by 51:
p$ncensor.plot <- p$ncensor.plot
scale_y_continuous(breaks = seq(12))
scale_x_continuous(breaks = c(0, 13, 26, 39, 51), limits = c(NA, 51))
p