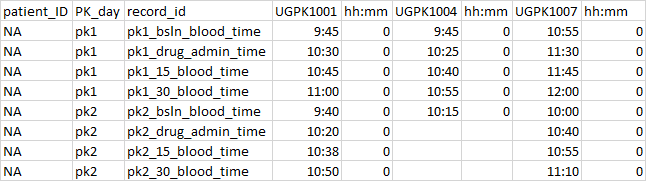
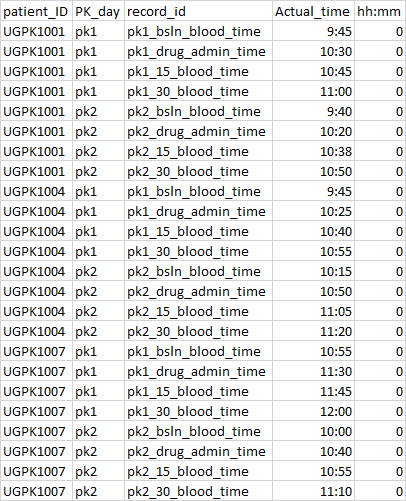
How can I rearrange columns and rows while also repeating column names for the length of the column?
Is there a way to make a data frame that looks like this...
Into this...
Edit:
Starting data...
df <- structure(list(patient_ID = c(NA, NA, NA, NA, NA, NA, NA, NA),
PK_day = c("pk1", "pk1", "pk1", "pk1", "pk2", "pk2", "pk2",
"pk2"), record_id = c("pk1_bsin_blood_time", "pk1_drug_admin_time",
"pk1_15_blood_time", "pk1_30_blood_time", "pk2_bsIn_blood_time",
"pk2_drug_admin_time", "pk2_15_blood_time", "pk2_30_blood_time"
), UGPK1001 = c("9:45", "10:30", "10:45", "11:00", "9:40",
"10:20", "10:38", "10:50"), hh.mm = c(0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L), UGPK1004 = c("9:45", "10:25", "10:40", "10:55",
"10:15", NA, NA, NA), hh.mm.1 = c(0L, 0L, 0L, 0L, 0L, NA,
NA, NA), UGPK1007 = c("10:55", "11:30", "11:45", "12:00",
"10:00", "10:40", "10:55", "11:10"), hh.mm.2 = c(0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L)), class = "data.frame", row.names = c(NA,
-8L))
CodePudding user response:
We could do it this way.
It remains unclear with the NA time which I removed with filter(!is.na(Actual_time))
If we know how they are generated in the long format then just remove the filter and add to the dataset!
library(dplyr)
library(tidyr)
df %>%
select(-patient_ID) %>%
pivot_longer(
c(UGPK1001, UGPK1004, UGPK1007),
names_to = "patient_ID",
values_to = "Actual_time"
) %>%
arrange(patient_ID) %>%
filter(!is.na(Actual_time)) %>%
select(patient_ID, PK_day, record_id, Actual_time, `hh:mm`=hh.mm)
patient_ID PK_day record_id Actual_time hh:mm
1 UGPK1001 pk1 pk1_bsin_blood_time 9:45 0
2 UGPK1001 pk1 pk1_drug_admin_time 10:30 0
3 UGPK1001 pk1 pk1_15_blood_time 10:45 0
4 UGPK1001 pk1 pk1_30_blood_time 11:00 0
5 UGPK1001 pk2 pk2_bsIn_blood_time 9:40 0
6 UGPK1001 pk2 pk2_drug_admin_time 10:20 0
7 UGPK1001 pk2 pk2_15_blood_time 10:38 0
8 UGPK1001 pk2 pk2_30_blood_time 10:50 0
9 UGPK1004 pk1 pk1_bsin_blood_time 9:45 0
10 UGPK1004 pk1 pk1_drug_admin_time 10:25 0
11 UGPK1004 pk1 pk1_15_blood_time 10:40 0
12 UGPK1004 pk1 pk1_30_blood_time 10:55 0
13 UGPK1004 pk2 pk2_bsIn_blood_time 10:15 0
14 UGPK1007 pk1 pk1_bsin_blood_time 10:55 0
15 UGPK1007 pk1 pk1_drug_admin_time 11:30 0
16 UGPK1007 pk1 pk1_15_blood_time 11:45 0
17 UGPK1007 pk1 pk1_30_blood_time 12:00 0
18 UGPK1007 pk2 pk2_bsIn_blood_time 10:00 0
19 UGPK1007 pk2 pk2_drug_admin_time 10:40 0
20 UGPK1007 pk2 pk2_15_blood_time 10:55 0
21 UGPK1007 pk2 pk2_30_blood_time 11:10 0
data:
df <- structure(list(patient_ID = c(NA, NA, NA, NA, NA, NA, NA, NA),
PK_day = c("pk1", "pk1", "pk1", "pk1", "pk2", "pk2", "pk2",
"pk2"), record_id = c("pk1_bsin_blood_time", "pk1_drug_admin_time",
"pk1_15_blood_time", "pk1_30_blood_time", "pk2_bsIn_blood_time",
"pk2_drug_admin_time", "pk2_15_blood_time", "pk2_30_blood_time"
), UGPK1001 = c("9:45", "10:30", "10:45", "11:00", "9:40",
"10:20", "10:38", "10:50"), hh.mm = c(0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L), UGPK1004 = c("9:45", "10:25", "10:40", "10:55",
"10:15", NA, NA, NA), hh.mm.1 = c(0L, 0L, 0L, 0L, 0L, NA,
NA, NA), UGPK1007 = c("10:55", "11:30", "11:45", "12:00",
"10:00", "10:40", "10:55", "11:10"), hh.mm.2 = c(0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L)), class = "data.frame", row.names = c(NA,
-8L))