my data:
data <- structure(list(col1 = c(10L, 20L, 30L, 40L, 50L, 60L, 70L, 80L,
90L, 100L, 20L, 30L, 40L, 50L, 60L), col2 = c(20L, 30L, 40L,
50L, 60L, 70L, 80L, 90L, 100L, 110L, 30L, 40L, 50L, 60L, 70L),
col3 = c(30L, 40L, 50L, 60L, 70L, 80L, 90L, 100L, 110L, 1120L,
40L, 50L, 60L, 70L, 80L), col4 = c(40L, 50L, 60L, 70L, 80L,
90L, 100L, 110L, 120L, 130L, 50L, 60L, 70L, 80L, 90L), group1 = c(1L,
1L, NA, 1L, NA, NA, 1L, NA, 1L, NA, 2L, 2L, 2L, 2L, 2L),
group2 = c(NA, NA, 1L, NA, 1L, 1L, NA, 1L, NA, 1L, 2L, 2L,
2L, 2L, 2L)), class = "data.frame", row.names = c(NA, -15L
))
I want to get one from several columns in which the columns go one after the other, first the first group, and then the second.
what I want to get:
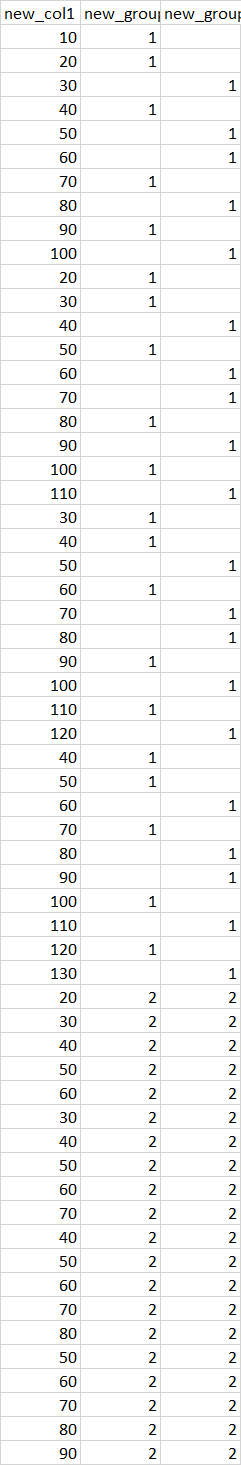
structure(list(new_col1 = c(10L, 20L, 30L, 40L, 50L, 60L, 70L,
80L, 90L, 100L, 20L, 30L, 40L, 50L, 60L, 70L, 80L, 90L, 100L,
110L, 30L, 40L, 50L, 60L, 70L, 80L, 90L, 100L, 110L, 120L, 40L,
50L, 60L, 70L, 80L, 90L, 100L, 110L, 120L, 130L, 20L, 30L, 40L,
50L, 60L, 30L, 40L, 50L, 60L, 70L, 40L, 50L, 60L, 70L, 80L, 50L,
60L, 70L, 80L, 90L), new_group1 = c(1L, 1L, NA, 1L, NA, NA, 1L,
NA, 1L, NA, 1L, 1L, NA, 1L, NA, NA, 1L, NA, 1L, NA, 1L, 1L, NA,
1L, NA, NA, 1L, NA, 1L, NA, 1L, 1L, NA, 1L, NA, NA, 1L, NA, 1L,
NA, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L), new_group2 = c(NA, NA, 1L, NA, 1L, 1L, NA,
1L, NA, 1L, NA, NA, 1L, NA, 1L, 1L, NA, 1L, NA, 1L, NA, NA, 1L,
NA, 1L, 1L, NA, 1L, NA, 1L, NA, NA, 1L, NA, 1L, 1L, NA, 1L, NA,
1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L)), class = "data.frame", row.names = c(NA,
-60L))
CodePudding user response:
Another possible solution, based on tidyr::pivot_longer:
library(tidyverse)
data %>%
pivot_longer(starts_with("col"), names_to = NULL, values_to = "new_col")
#> # A tibble: 60 x 3
#> group1 group2 new_col
#> <int> <int> <int>
#> 1 1 NA 10
#> 2 1 NA 20
#> 3 1 NA 30
#> 4 1 NA 40
#> 5 1 NA 20
#> 6 1 NA 30
#> 7 1 NA 40
#> 8 1 NA 50
#> 9 NA 1 30
#> 10 NA 1 40
#> # ... with 50 more rows
CodePudding user response:
library(dplyr)
library(tidyr)
data %>%
gather(var, new_col, -group1, -group2) %>%
select(new_col, new_group1 = group1, new_group2 = group2)
CodePudding user response:
Here's another option using data.table:
library(data.table)
dt <- as.data.table(data)
output <-
setnames(melt(dt, id.vars = c('group1', 'group2'))[, c(4, 1:2)],
new = c("new_col1", "new_group1", "new_group2"))
Output
output
new_col1 new_group1 new_group2
1: 10 1 NA
2: 20 1 NA
3: 30 NA 1
4: 40 1 NA
5: 50 NA 1
6: 60 NA 1
7: 70 1 NA
8: 80 NA 1
9: 90 1 NA
10: 100 NA 1
11: 20 2 2
12: 30 2 2
13: 40 2 2
14: 50 2 2
15: 60 2 2
16: 20 1 NA
17: 30 1 NA
18: 40 NA 1
19: 50 1 NA
20: 60 NA 1
21: 70 NA 1
22: 80 1 NA
23: 90 NA 1
24: 100 1 NA
25: 110 NA 1
26: 30 2 2
27: 40 2 2
28: 50 2 2
29: 60 2 2
30: 70 2 2
31: 30 1 NA
32: 40 1 NA
33: 50 NA 1
34: 60 1 NA
35: 70 NA 1
36: 80 NA 1
37: 90 1 NA
38: 100 NA 1
39: 110 1 NA
40: 1120 NA 1
41: 40 2 2
42: 50 2 2
43: 60 2 2
44: 70 2 2
45: 80 2 2
46: 40 1 NA
47: 50 1 NA
48: 60 NA 1
49: 70 1 NA
50: 80 NA 1
51: 90 NA 1
52: 100 1 NA
53: 110 NA 1
54: 120 1 NA
55: 130 NA 1
56: 50 2 2
57: 60 2 2
58: 70 2 2
59: 80 2 2
60: 90 2 2