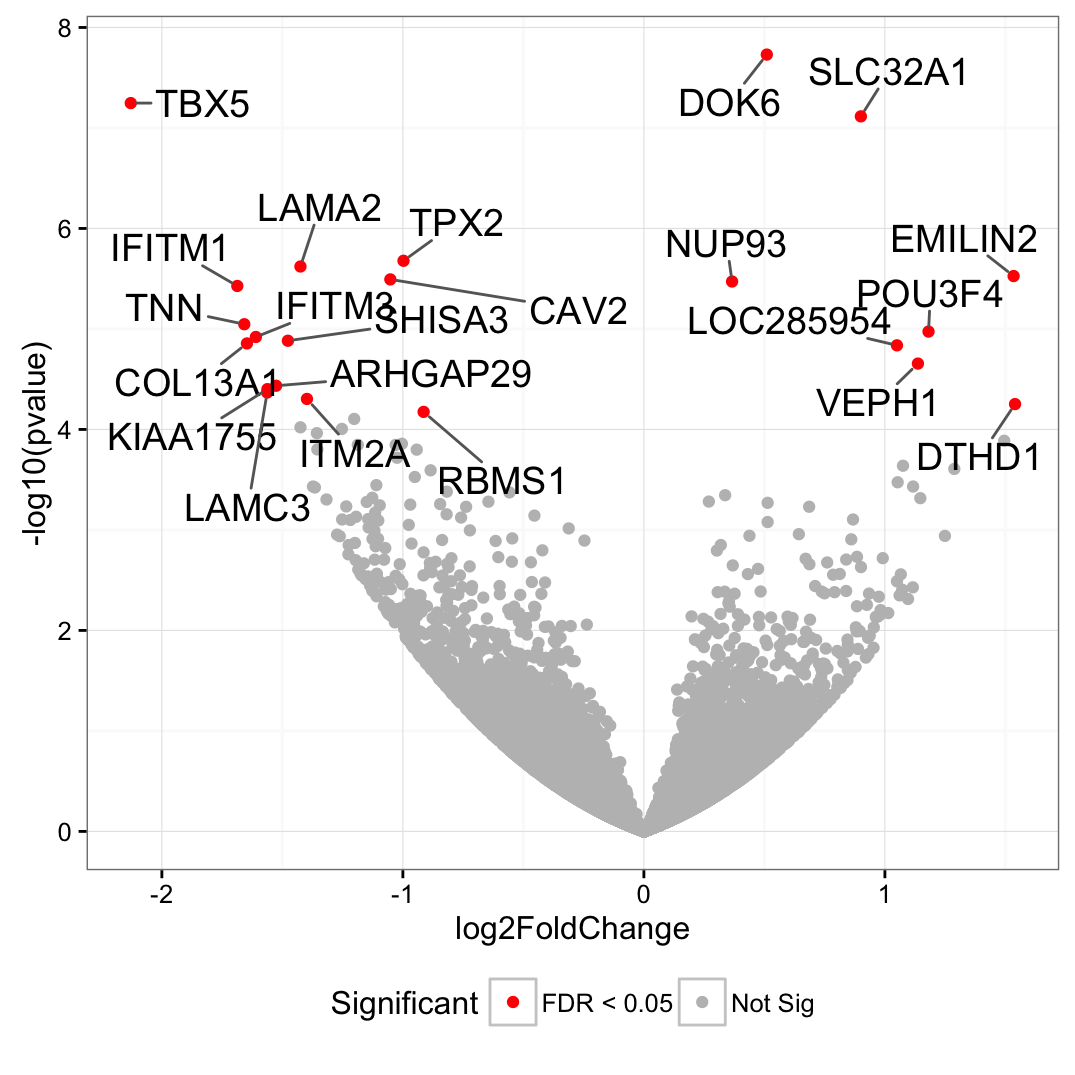
Consider the following example where a scatter is made and only the "significant" point are colored and labelled.
genes <- read.table("https://gist.githubusercontent.com/stephenturner/806e31fce55a8b7175af/raw/1a507c4c3f9f1baaa3a69187223ff3d3050628d4/results.txt", header = TRUE)
genes$Significant <- ifelse(genes$padj < 0.05, "FDR < 0.05", "Not Sig")
ggplot(genes, aes(x = log2FoldChange, y = -log10(pvalue)))
geom_point(aes(color = Significant))
scale_color_manual(values = c("red", "grey"))
theme_bw(base_size = 12) theme(legend.position = "bottom")
geom_text_repel(
data = subset(genes, padj < 0.05),
aes(label = Gene),
size = 5,
box.padding = unit(0.35, "lines"),
point.padding = unit(0.3, "lines")
)
Now imagine that the labels are actually acronyms and that they have a real full-length name (e.g., "DOK6" is the acronym for "Duo Ocarino Kayne 6"). Would it be possible to a legend to the plot where the keys are the labels used on the plot, and the entries are the full-length name of the labels ?
CodePudding user response:
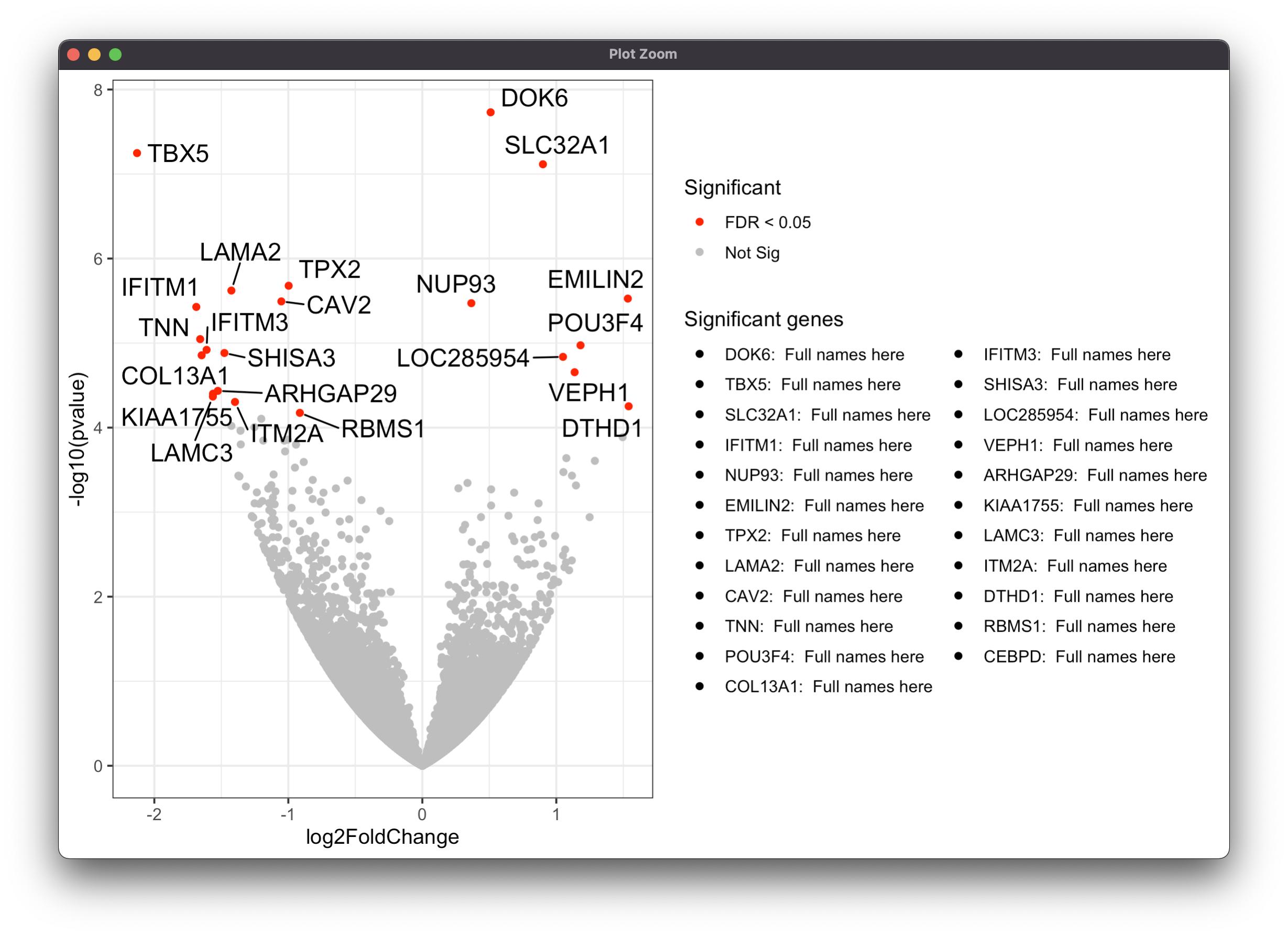
First, I added Gene2 for another legend which only shows significant Gene.\
Next, Gene2 was added on the aes as a fill. (Only color would affect the color of points on geom_point).\
Finally, scale_fill_discrete was added for the second legend. All you need to do is just annotate the full-length name column at Full names here.
genes$Gene2 <-ifelse(genes$padj<0.05, genes$Gene, NA)
ggplot(genes, aes(x = log2FoldChange, y = -log10(pvalue),
fill=Gene2))
geom_point(aes(color = Significant))
scale_color_manual(values = c("red", "grey"))
theme_bw(base_size = 12) theme(legend.position = "bottom")
geom_text_repel(
data = subset(genes, padj < 0.05),
aes(label = Gene),
size = 5,
box.padding = unit(0.35, "lines"),
point.padding = unit(0.3, "lines")
)
scale_fill_discrete(labels=paste0(genes$Gene,': ',' Full names here'),
name='Significant genes')
theme(legend.position = 'right')