I have a huge dataset with over 2 million obs and 100 columns. There are 9 columns that contain ICD-10 disease codes that each of them start with a different letter.
For example:
icd1 <- c("O230", "B540", "D990", "Y555", "E980", "J777", "P090", "Q090", "R433")
icd2 <- c("O230", "B540", "D990", "Y555", "E980", "J777", "P090", "Q090", "Z433")
icd3 <- c("X230", "B540", "D990", "Y555", "E980", "J777", "P090", "Q090", "Z433")
data <- as.data.frame(rbind(icd1, icd2, icd3))
What I need to do is create a new column called "ICD_Z" that checks every one of those 9 ICD-10 columns codes to see if any of them starts with the letter Z, so if that's the case, the column I created would receive 1, or else, 0. But it only can receive 1 if all those 9 columns don't have an ICD-10 code that starts with the letter O.
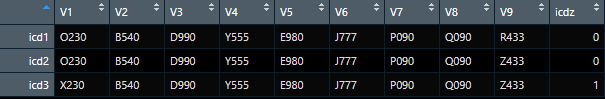
So my output would look like this:
How can I do that?
CodePudding user response:
You could use if_any and if_all:
data %>%
mutate(ICD_Z = if_any(V1:V9, ~grepl('^Z', .)) *
if_all(V1:V9, ~!grepl('^O', .)))
V1 V2 V3 V4 V5 V6 V7 V8 V9 ICD_Z
icd1 O230 B540 D990 Y555 E980 J777 P090 Q090 R433 0
icd2 O230 B540 D990 Y555 E980 J777 P090 Q090 Z433 0
icd3 X230 B540 D990 Y555 E980 J777 P090 Q090 Z433 1
Edit:
You can use a variable to store the positions of the columns in question:
nms <- c(1,2,3,5,7,8)
data %>%
mutate(ICD_Z = if_any(all_of(nms), ~grepl('^Z', .)) *
if_all(all_of(nms), ~!grepl('^O', .)))
CodePudding user response:
With a combination of rowwise() and c_across() it's very straightforward:
library(dplyr)
icd1 <- c("O230", "B540", "D990", "Y555", "E980", "J777", "P090", "Q090", "R433")
icd2 <- c("O230", "B540", "D990", "Y555", "E980", "J777", "P090", "Q090", "Z433")
icd3 <- c("X230", "B540", "D990", "Y555", "E980", "J777", "P090", "Q090", "Z433")
data <- as.data.frame(rbind(icd1, icd2, icd3))
data %>%
rowwise() %>%
mutate(has_z = any(startsWith(c_across(V1:V9), "Z")),
has_o = any(startsWith(c_across(V1:V9), "O")),
icdz = as.numeric(has_z & !has_o)
) %>%
select(-has_z, -has_o)
#> # A tibble: 3 × 10
#> # Rowwise:
#> V1 V2 V3 V4 V5 V6 V7 V8 V9 icdz
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 O230 B540 D990 Y555 E980 J777 P090 Q090 R433 0
#> 2 O230 B540 D990 Y555 E980 J777 P090 Q090 Z433 0
#> 3 X230 B540 D990 Y555 E980 J777 P090 Q090 Z433 1
Created on 2022-05-25 by the reprex package (v2.0.1)
CodePudding user response:
Update: an even faster approach:
icd_cols = paste0("V",1:9)
f <- function(v) !any(v=="O") & any(v=="Z")
setDT(data)
set(data, j="icdz", value= apply(data[,..icd_cols],1,\(i) f(substr(i,1,1))))
Original Solution
It is possible with your large dataset you will have some speed up with this approach, that uses data.table. On the small 3 row example, I found informally this to be about 5 times faster.
- identify the list of 9 ICD columns; I've used a simple construction here to get V1:V9, but you could use
icd_cols = colnames(data)[c(20,21,37:45)] - create a small function that evaluates a vector of first letters
- set the original frame as data.table
- melt and apply the function by id, and assign the result to the original frame
icd_cols = paste0("V",1:9)
f <- function(v) !any(v=="O") & any(v=="Z")
setDT(data)
data[, icdz:=(melt(data[,id:=.I],"id",icd_cols)[,f(substr(value,1,1)), by=id]$V1)][id:=NULL]
Output:
V1 V2 V3 V4 V5 V6 V7 V8 V9 icdz
1: O230 B540 D990 Y555 E980 J777 P090 Q090 R433 FALSE
2: O230 B540 D990 Y555 E980 J777 P090 Q090 Z433 FALSE
3: X230 B540 D990 Y555 E980 J777 P090 Q090 Z433 TRUE