Although, the figure is rather small, one can see that several points/events (lightred) do not lie on colored line segments and vice a versa. Where does this deviation come from? Is there a way to improve my procedure?
Why I am doing this? Overall goal:
- Attach point weights as edge weights to respective line segments;
- then plot line segments color or weight scaled (even better invent sth. like a heat map).
# Smallest reproducible example
library(spatstat)
data("chicago")
N <- npoints(chicago)
# create some weights, later useful
marks(chicago) <- rnorm(N, 5, 2)
# find to each point nearest line segment
idx <- nearestsegment(as.ppp(chicago), as.psp(chicago))
# not to my suprise this is quite similar to:
# (i) retrieve network
lns <- sf::st_as_sf(maptools::as.SpatialLines.psp(
spatstat.geom::as.psp(chicago)))
# (ii) retrieve event coords
events <- data.frame(xcoords = chicago[["data"]]$x,
ycoords = chicago[["data"]]$y)
# convert
pnts <- sf::st_as_sf(events, coords = c("xcoords", "ycoords"))
# get nearest feature
idx2 <- sf::st_nearest_feature(pnts, lns)
# one deviation
which(idx != idx2)
#> [1] 1
idx[1]
#> [1] 37
idx2[2]
#> [1] 54
# my overall goal is to transmit the point weights to the
# respective line segments. With those edge weights I would like to
# plot some maps, therefore:
# (I) create igraph object from chicago:
library(igraph)
edges <- cbind(chicago[["domain"]][["from"]], chicago[["domain"]][["to"]])
chicago_net <- graph_from_edgelist(edges)
adjm <- as.matrix(as_adjacency_matrix(chicago_net))
g <- graph_from_adjacency_matrix(adjm, 'undirected')
vertex_attr(g, name = 'x') <- chicago[["domain"]]$vertices[["x"]]
vertex_attr(g, name = 'y') <- chicago[["domain"]]$vertices[["y"]]
# (II) add point weights as edge weights to graph according to idx:
weights <- c(rep(NA, nrow(chicago[["domain"]]$lines[["ends"]])))
weights[idx2] <- chicago[["data"]]$marks
E(g)$weight <- weights
# finally plot g via ggraph [same result with igraph.plot()]
library(ggraph)
ggraph(g, x = vertex_attr(g)$x, y = vertex_attr(g)$y)
geom_edge_link0(aes(color = weight))
# sth. practical from here:
# https://ggraph.data-imaginist.com/reference/scale_edge_colour.html
scale_edge_colour_viridis()
geom_point(data = events, mapping = aes(x = xcoords, y = ycoords),
alpha = 0.3, size = 2, shape = 20, col = "red")
theme_void()
gives
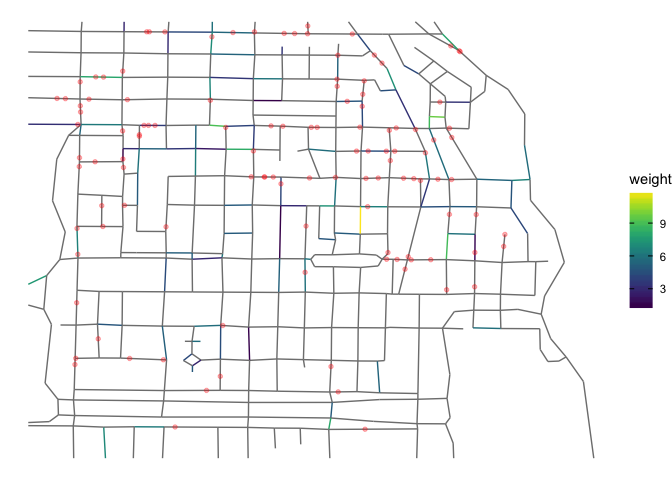
Created on 2022-05-25 by the reprex package (v2.0.1)
FYI ?nearestsegment's description is as follows
Given a point pattern and a line segment pattern, this function finds the nearest line segment for each point.
Sooner or later, everything I do with {spatstat} works as it should, hence I think I am doing a great mistake.
CodePudding user response:
remove adjacency matrix calculation; it reorders segments of the graph
correct g assignment:
g <- graph_from_edgelist(edges)
