I am new to mapping data with R and I would like some help for a complex representation. I will try to be as clear as possible :)
I have a dataset that counts observations over time since 1950 per day in Sweden. Every line is an observation, with a latitude, longitude, julian day, date & year informations. I have divided Sweden in three groups of latitude (1 is south, 2 is middle, 3 is north). I care only about the latitude information, so that the longitude can be turned into the same value for every point if needed.
I would like to map the change over time in terms of density of observations in these three groups. To do so, I would like to represent the change at different key years of my dataset: 1950/1975/2000/2021, so I need to create multiple maps. Plus, I would like for each year a map of the density of cumulated observations for the 15 first days of February/15 last days of March/15 first days of April & 15 last days of May ; so that the total of maps would be 4*4 = 16. Ideally, the change would be symbolized by a colour gradient (the darker the more observations). But I don't mind other suggestions if it is not appropriate.
Random sample of my big dataset:
> dput(df[sample(nrow(df), 50),])
structure(list(lat = c("65", "64", "65", "59", "59", "57", "57",
"68", "67", "63", "60", "61", "65", "59", "56", "65", "59", "57",
"55", "59", "56", "56", "59", "60", "59", "55", "59", "59", "57",
"55", "56", "57", "65", "59", "63", "59", "56", "59", "56", "56",
"57", "63", "58", "59", "63", "61", "55", "58", "66", "57"),
long = c("21", "17", "21", "14", "14", "13", "12", "18",
"18", "20", "16", "14", "17", "16", "12", "16", "15", "14",
"12", "17", "12", "16", "18", "14", "14", "14", "18", "17",
"12", "13", "12", "12", "21", "13", "19", "16", "12", "18",
"16", "12", "12", "18", "12", "17", "20", "17", "12", "13",
"19", "12"), date = c("2009-03-29", "2006-04-06", "2019-03-31",
"2006-04-04", "1975-04-13", "2014-02-05", "1996-04-02", "2021-04-08",
"1995-04-12", "2004-04-12", "2018-04-07", "2021-03-28", "1988-04-01",
"2002-03-17", "2015-03-12", "2019-04-05", "2016-03-19", "2021-04-03",
"2014-02-08", "2015-03-13", "2021-03-09", "2005-02-07", "2013-03-31",
"1989-03-23", "1989-03-27", "2015-01-21", "2011-04-04", "2018-03-26",
"1987-03-23", "2011-01-31", "2014-02-09", "2004-01-17", "2012-04-20",
"2017-03-07", "2005-04-02", "2017-01-28", "2016-03-19", "1984-03-30",
"2005-01-29", "2021-03-06", "2008-02-03", "2017-03-22", "2019-03-10",
"2010-01-17", "2009-04-10", "2016-01-23", "2019-03-01", "2006-03-04",
"2014-04-23", "2009-03-15"), julian_day = c("88", "96", "90",
"94", "103", "36", "93", "98", "102", "103", "97", "87",
"92", "76", "71", "95", "79", "93", "39", "72", "68", "38",
"90", "82", "86", "21", "94", "85", "82", "31", "40", "17",
"111", "66", "92", "28", "79", "90", "29", "65", "34", "81",
"69", "17", "100", "23", "60", "63", "113", "74"), year = c(2009L,
2006L, 2019L, 2006L, 1975L, 2014L, 1996L, 2021L, 1995L, 2004L,
2018L, 2021L, 1988L, 2002L, 2015L, 2019L, 2016L, 2021L, 2014L,
2015L, 2021L, 2005L, 2013L, 1989L, 1989L, 2015L, 2011L, 2018L,
1987L, 2011L, 2014L, 2004L, 2012L, 2017L, 2005L, 2017L, 2016L,
1984L, 2005L, 2021L, 2008L, 2017L, 2019L, 2010L, 2009L, 2016L,
2019L, 2006L, 2014L, 2009L), lat_grouped = c("3", "2", "3",
"1", "1", "1", "1", "3", "3", "2", "2", "2", "3", "1", "1",
"3", "1", "1", "1", "1", "1", "1", "1", "2", "1", "1", "1",
"1", "1", "1", "1", "1", "3", "1", "2", "1", "1", "1", "1",
"1", "1", "2", "1", "1", "2", "2", "1", "1", "3", "1")), row.names = c(22330L,
15394L, 44863L, 15258L, 1481L, 31695L, 6399L, 52043L, 6111L,
11508L, 42184L, 51391L, 4308L, 8764L, 34675L, 45080L, 37042L,
51743L, 31717L, 34723L, 50514L, 11892L, 30527L, 4572L, 4608L,
33744L, 26476L, 41366L, 4006L, 25265L, 31741L, 10122L, 29059L,
38340L, 12787L, 37827L, 37061L, 3029L, 11762L, 50464L, 18114L,
39026L, 43835L, 23081L, 22811L, 36179L, 43641L, 13743L, 33608L,
21917L), class = "data.frame")
I have managed to create the base layer following some guidelines found on the internet, but I have no clue on how to keep going and I am being confused by all the different methods that I have not managed to make working.
library(ggplot2)
library(gganimate)
library(gifski)
library(maps)
library(sf)
library(rgdal)
#map source: https://www.geoboundaries.org/data/1_3_3/zip/shapefile/
wd = "C:/Users/HP/Desktop/SWE_ADM0"
sweden <- readOGR(paste0(wd, "/SWE_ADM0.shp"), layer = "SWE_ADM0")
plot(sweden)
#To use the imported shapefile in ggmap, we need the fortify() function of the ggplot2 package.
sweden_fort <- ggplot2::fortify(sweden)
base_map <- ggplot(data = sweden_fort, mapping = aes(x=long, y=lat, group=group))
geom_polygon(color = "black", fill = "white")
coord_quickmap()
theme_void()
base_map
I hope that someone will be able to give me a hand, if something is not clear or info is missing, I can edit my post :)
Many thanks.
CodePudding user response:
If I were you I'd use sf objects, i.e. read in the sweden map with st_read() rather than using readOGR() directly and then using fortify(). This will let you use geom_sf() rather than geom_polygon(). In addition you should simplify the sweden shapefile you are using. The one you point to is very detailed, i.e. very many lines. If you try to use it in an animation, it will take hours and hours to render. You can simplify it quite a lot without loss of relevant detail for your plot. Create the df as an sf object too---one consisting of long/lat points rather than lines---and then you should be good to go.
So, using your df above and the map of Sweden you pointed to,
library(tidyverse)
library(sf)
library(here)
#map source: https://www.geoboundaries.org/data/1_3_3/zip/shapefile/
## Simplify the map for quicker rendering
sweden <- st_read(here("data", "SWE_ADM0", "SWE_ADM0.shp"),
layer = "SWE_ADM0") |>
st_simplify(dTolerance = 1e3)
#> Reading layer `SWE_ADM0' from data source `scratch/data/SWE_ADM0/SWE_ADM0.shp' using driver `ESRI Shapefile'
#> Simple feature collection with 1 feature and 8 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: 10.98139 ymin: 55.33695 xmax: 24.16663 ymax: 69.05997
#> Geodetic CRS: WGS 84
df <- structure(list(lat = c("65", "64", "65", "59", "59", "57", "57",
"68", "67", "63", "60", "61", "65", "59", "56", "65", "59", "57",
"55", "59", "56", "56", "59", "60", "59", "55", "59", "59", "57",
"55", "56", "57", "65", "59", "63", "59", "56", "59", "56", "56",
"57", "63", "58", "59", "63", "61", "55", "58", "66", "57"),
long = c("21", "17", "21", "14", "14", "13", "12", "18",
"18", "20", "16", "14", "17", "16", "12", "16", "15", "14",
"12", "17", "12", "16", "18", "14", "14", "14", "18", "17",
"12", "13", "12", "12", "21", "13", "19", "16", "12", "18",
"16", "12", "12", "18", "12", "17", "20", "17", "12", "13",
"19", "12"), date = c("2009-03-29", "2006-04-06", "2019-03-31",
"2006-04-04", "1975-04-13", "2014-02-05", "1996-04-02", "2021-04-08",
"1995-04-12", "2004-04-12", "2018-04-07", "2021-03-28", "1988-04-01",
"2002-03-17", "2015-03-12", "2019-04-05", "2016-03-19", "2021-04-03",
"2014-02-08", "2015-03-13", "2021-03-09", "2005-02-07", "2013-03-31",
"1989-03-23", "1989-03-27", "2015-01-21", "2011-04-04", "2018-03-26",
"1987-03-23", "2011-01-31", "2014-02-09", "2004-01-17", "2012-04-20",
"2017-03-07", "2005-04-02", "2017-01-28", "2016-03-19", "1984-03-30",
"2005-01-29", "2021-03-06", "2008-02-03", "2017-03-22", "2019-03-10",
"2010-01-17", "2009-04-10", "2016-01-23", "2019-03-01", "2006-03-04",
"2014-04-23", "2009-03-15"), julian_day = c("88", "96", "90",
"94", "103", "36", "93", "98", "102", "103", "97", "87",
"92", "76", "71", "95", "79", "93", "39", "72", "68", "38",
"90", "82", "86", "21", "94", "85", "82", "31", "40", "17",
"111", "66", "92", "28", "79", "90", "29", "65", "34", "81",
"69", "17", "100", "23", "60", "63", "113", "74"), year = c(2009L,
2006L, 2019L, 2006L, 1975L, 2014L, 1996L, 2021L, 1995L, 2004L,
2018L, 2021L, 1988L, 2002L, 2015L, 2019L, 2016L, 2021L, 2014L,
2015L, 2021L, 2005L, 2013L, 1989L, 1989L, 2015L, 2011L, 2018L,
1987L, 2011L, 2014L, 2004L, 2012L, 2017L, 2005L, 2017L, 2016L,
1984L, 2005L, 2021L, 2008L, 2017L, 2019L, 2010L, 2009L, 2016L,
2019L, 2006L, 2014L, 2009L), lat_grouped = c("3", "2", "3",
"1", "1", "1", "1", "3", "3", "2", "2", "2", "3", "1", "1",
"3", "1", "1", "1", "1", "1", "1", "1", "2", "1", "1", "1",
"1", "1", "1", "1", "1", "3", "1", "2", "1", "1", "1", "1",
"1", "1", "2", "1", "1", "2", "2", "1", "1", "3", "1")), row.names = c(22330L,
15394L, 44863L, 15258L, 1481L, 31695L, 6399L, 52043L, 6111L,
11508L, 42184L, 51391L, 4308L, 8764L, 34675L, 45080L, 37042L,
51743L, 31717L, 34723L, 50514L, 11892L, 30527L, 4572L, 4608L,
33744L, 26476L, 41366L, 4006L, 25265L, 31741L, 10122L, 29059L,
38340L, 12787L, 37827L, 37061L, 3029L, 11762L, 50464L, 18114L,
39026L, 43835L, 23081L, 22811L, 36179L, 43641L, 13743L, 33608L,
21917L), class = "data.frame")
## Convert the given sample data to an `sf` object of points, setting
## the coordinate system to be the same as the `sweden` map
df <- df |>
mutate(id = 1:nrow(df),
date = lubridate::ymd(date),
year = factor(lubridate::year(date))) |>
st_as_sf(coords = c("long", "lat"), crs = 4326)
# Subset the data to the years to you want, and create the plot
df_selected <- df |>
filter(year %in% c(1975, 1989, 2016, 2021))
ggplot()
geom_sf(data = sweden)
geom_sf(data = df_selected,
mapping = aes(color = lat_grouped))
facet_grid(lat_grouped ~ year)
guides(color = "none")
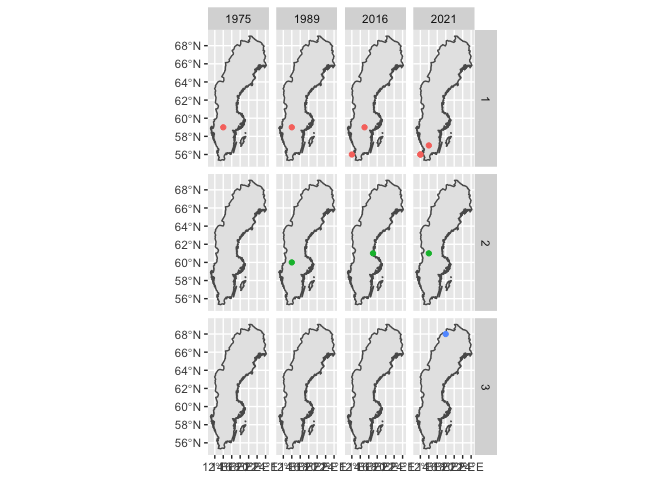
You can set e.g. theme_void() or map themes to get rid of the grid lines and so on.
Update: One last edit, just on the question of plotting densities. Once you calculate the cumulative data, you could for example overlay your map with 2D kernel density estimates. For example, here is a very rough first cut at that, faceted by latitude group.
ggplot()
geom_sf(data = sweden)
geom_density_2d_filled(data = df,
mapping = aes(x = map_dbl(geometry, ~.[1]),
y = map_dbl(geometry, ~.[2])),
alpha = 0.4)
facet_wrap(~ lat_grouped)
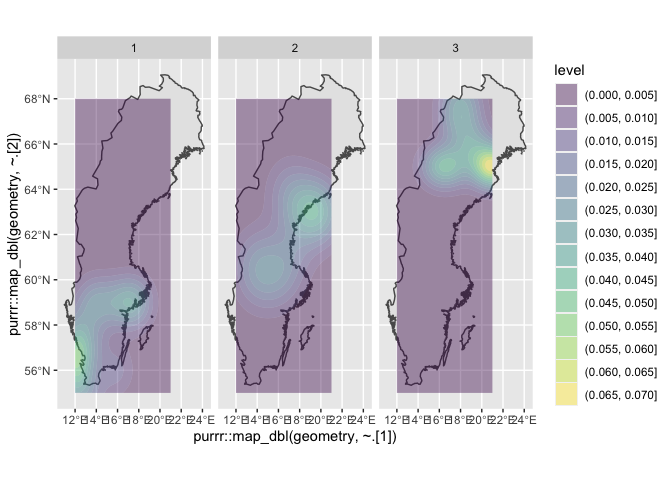
The map_dbl() function here (from the purrr package) is a way of reaching in to the geometry column of the df and extracting first the x (i.e. longitude) and then the y (i.e. latitude) data in order to give geom_density_2d() the coordinates it needs to calculate its estimates.
