input.txt: My actual input file has 5000000 lines
A B C D4.2 E 2022-05-31
A B C D4.2 E 2022-05-31
A B F D4.2 E 2022-05-07
A B C D4.2 E 2022-05-31
X B D E2.0 F 2022-05-30
X B Y D4.2 E 2022-05-06
data.txt : This is another file I need to refer in while loop.
A B C D4.2 E 2022-06-31
X B D E2.0 F 2022-07-30
Here's what I need to do
cat input.txt |while read foo bar tan ban can man
do
KEYVALUE=$(echo $4 |awk -F. '{print $1}')
END_DATE=`egrep -w '$1|${KEYVALUE}|$6' data.txt |awk '{print $5,$6}'`
echo $foo,$bar,$tan,$ban,$can,$man,${END_DATE}
done
Desired output:
A B C D4.2 E 2022-05-31 2022-06-31
A B C D4.2 E 2022-05-31 2022-06-31
A B F D4.2 E 2022-05-07 2022-06-31
A B C D4.2 E 2022-05-31 2022-06-31
X B D E2.0 F 2022-05-30 2022-07-30
X B Y D4.2 E 2022-05-06 2022-06-31
My major problem is the while loop takes more than an hour to complete going through 500000 input lines. How can I parallel process this since each line is independent of each other and the order of lines in output file doesn't matter. I've tried using GNU parallel based on few discussions. But none of them were helpful or maybe I am not sure how to implement it. I am using RHEL with BASH or KSH.
CodePudding user response:
If you achieve the development of a function to do whatever you need for each iteration, you could use 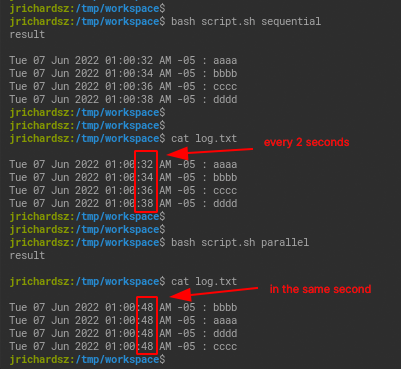
CodePudding user response:
Here is one potential solution:
cat script.awk
#!/usr/bin/awk -f
NR==FNR{
n=gsub("\.*","",$4)
a[n,$5]=$6; next
} (n,$5) in a {
print $0, a[n,$5]
}
cat input.txt | parallel --pipe -q ./script.awk data.txt -
A B C D4.2 E 2022-05-31 2022-06-31
A B C D4.2 E 2022-05-31 2022-06-31
A B F D4.2 E 2022-05-07 2022-06-31
A B C D4.2 E 2022-05-31 2022-06-31
X B D E2.0 F 2022-05-30 2022-07-30
X B Y D4.2 E 2022-05-06 2022-06-31
It should be relatively fast. You can tweak the parallel command (e.g. use --pipepart instead of --pipe) to increase performance depending on your parameters (i.e. size of each file, number of available cores, etc).
Edit
Rough benchmarking suggests it will be significantly faster:
# Copy input.txt many times
for f in {1..100}; do cat input.txt >> input.txt_2; done
for f in {1..1000}; do cat input.txt_2 >> input.txt_3; done
for f in {1..10}; do cat input.txt_3 >> input.txt_4; done
du -h input.txt_4
137M input.txt_4
wc -l input.txt_4
6000000 input.txt_4
time cat input.txt_4 | parallel --pipe -q ./script.awk data.txt - > output.txt
real 0m7.533s
user 0m22.085s
sys 0m4.494s
Took <10 seconds to process the 6M row input file. Does this solve your problem?
CodePudding user response:
Without parallel took 8 seconds for 5068056 lines
$ wc -l input.txt
5068056 input.txt
$ time awk 'NR==FNR{a[$4]=$6} NR!=FNR{print $0, a[$4]}' data.txt input.txt > output.txt
real 0m8.274s
user 0m5.397s
sys 0m2.869s
$ wc -l output.txt
5068056 output.txt
With parallel
time cat input.txt | parallel --pipe -q awk 'NR==FNR{a[$4]=$6; next} {print $0, a[$4]}' data.txt - > output.txt
real 0m3.319s
user 0m9.284s
sys 0m5.990s
Using split
inputfile=input.txt
outputfile=output.txt
data=data.txt
count=10
split -n l/$count $inputfile /tmp/input$$
for file in /tmp/input$$*; do
awk 'NR==FNR{a[$4]=$6; next} {print $0, a[$4]}' $data $file > ${file}.out &
done
wait
cat /tmp/input$$*.out > $outputfile
rm /tmp/input$$*
$ time ./split.sh
real 0m1.781s
user 0m7.244s
sys 0m1.536s
